DNA sequences of the crossover regions of EcoRI/XbaI-digested pUC19 and... | Download Scientific Diagram

Comparisons of the XbaI, NheI, and HindIII optical maps to sequence... | Download Scientific Diagram

Targeted Gene Knock In and Sequence Modulation Mediated by a Psoralen-linked Triplex-forming Oligonucleotide, - ScienceDirect
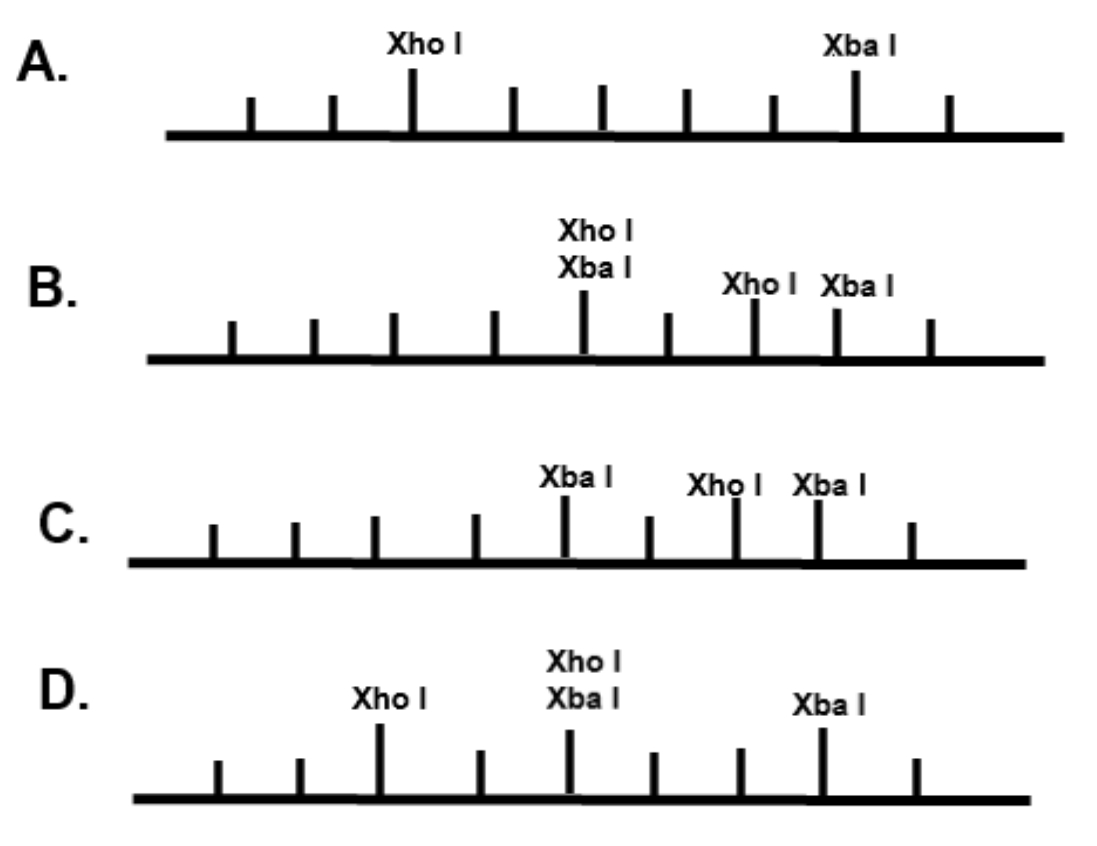
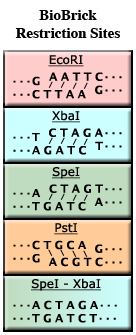
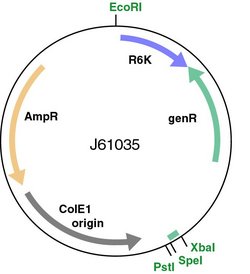
The restriction enzyme XhoI and XbaI were used to digest a linear fragment of DNA at a specific sequence such as that shown for EcoR1 (figure above). Use the information gained from
Silencing of HaAce1 gene by host-delivered artificial microRNA disrupts growth and development of Helicoverpa armigera | PLOS ONE
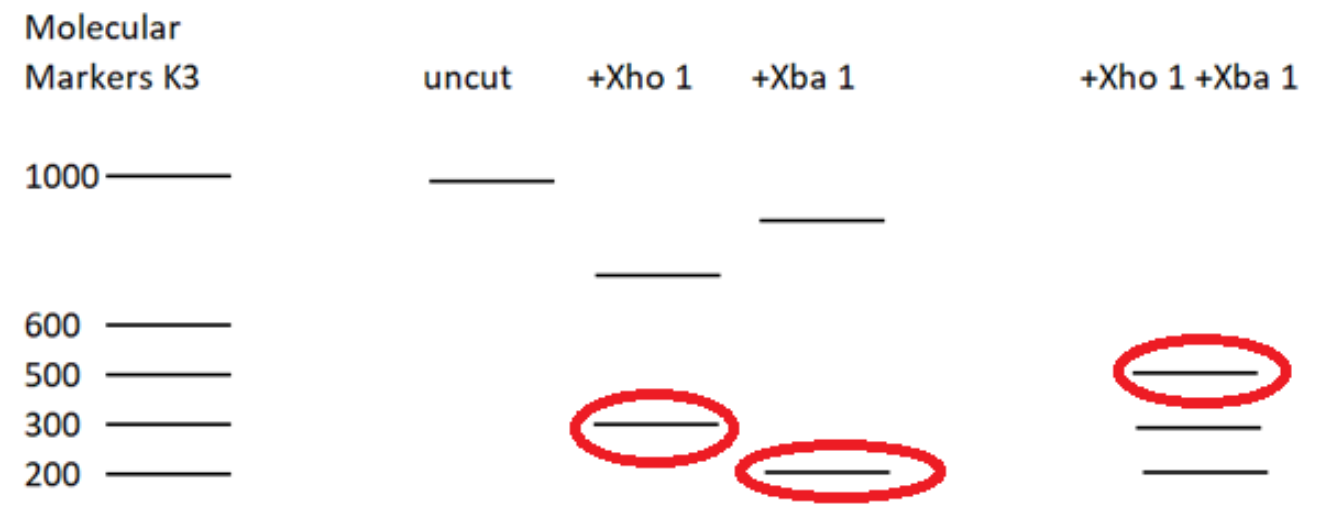
The restriction enzyme XhoI and XbaI were used to digest a linear fragment of DNA at a specific sequence such as that shown for EcoR1 (figure above). Use the information gained from