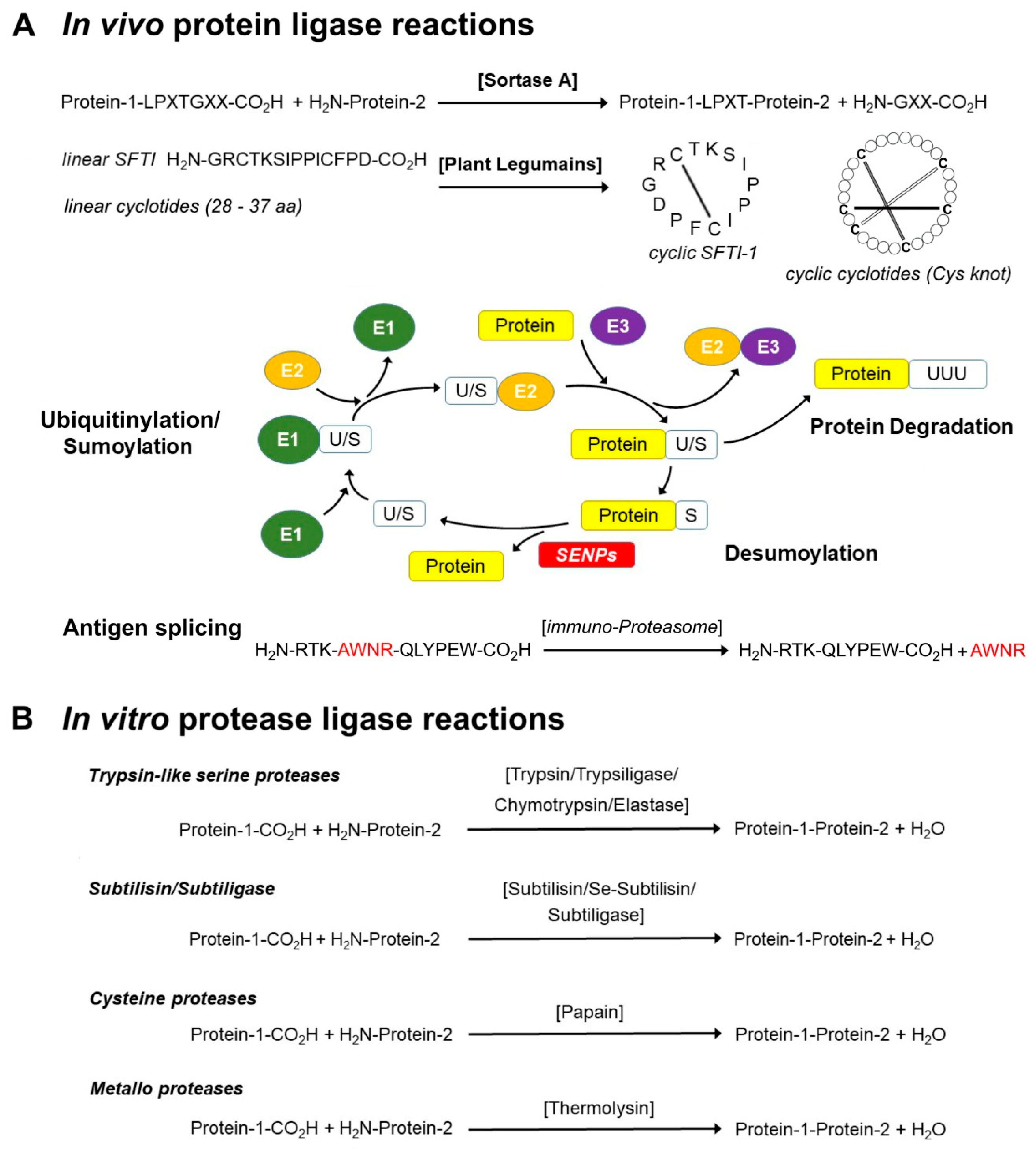
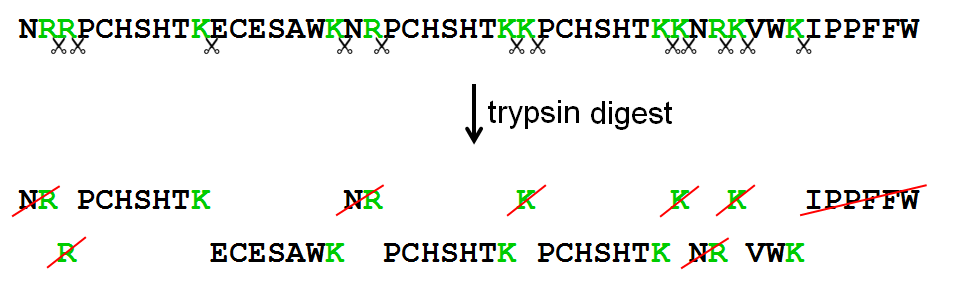
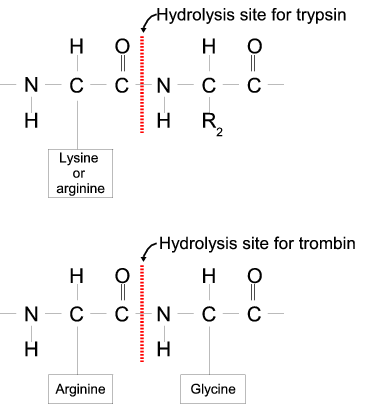
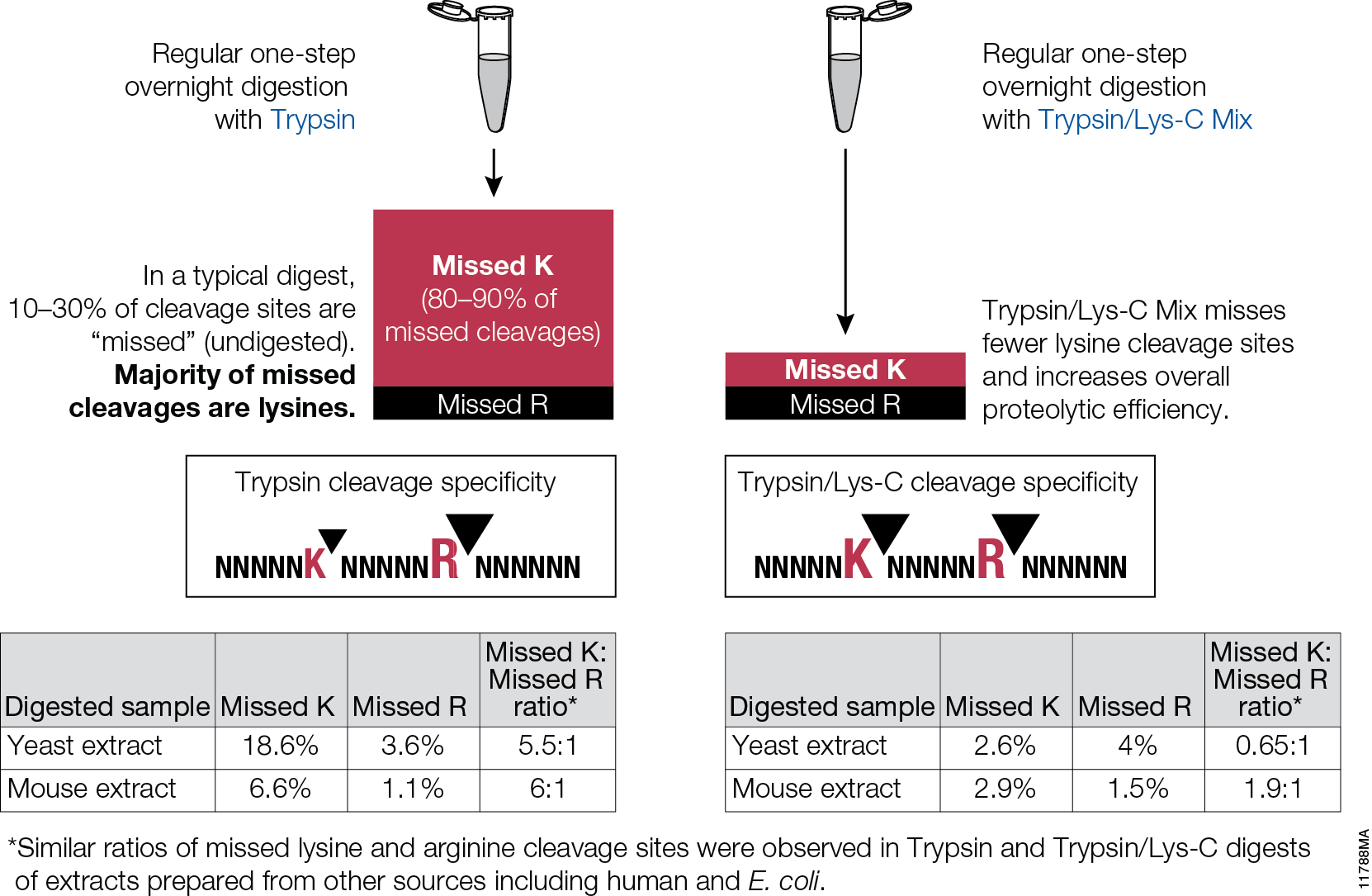
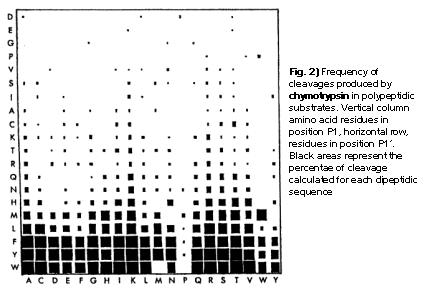
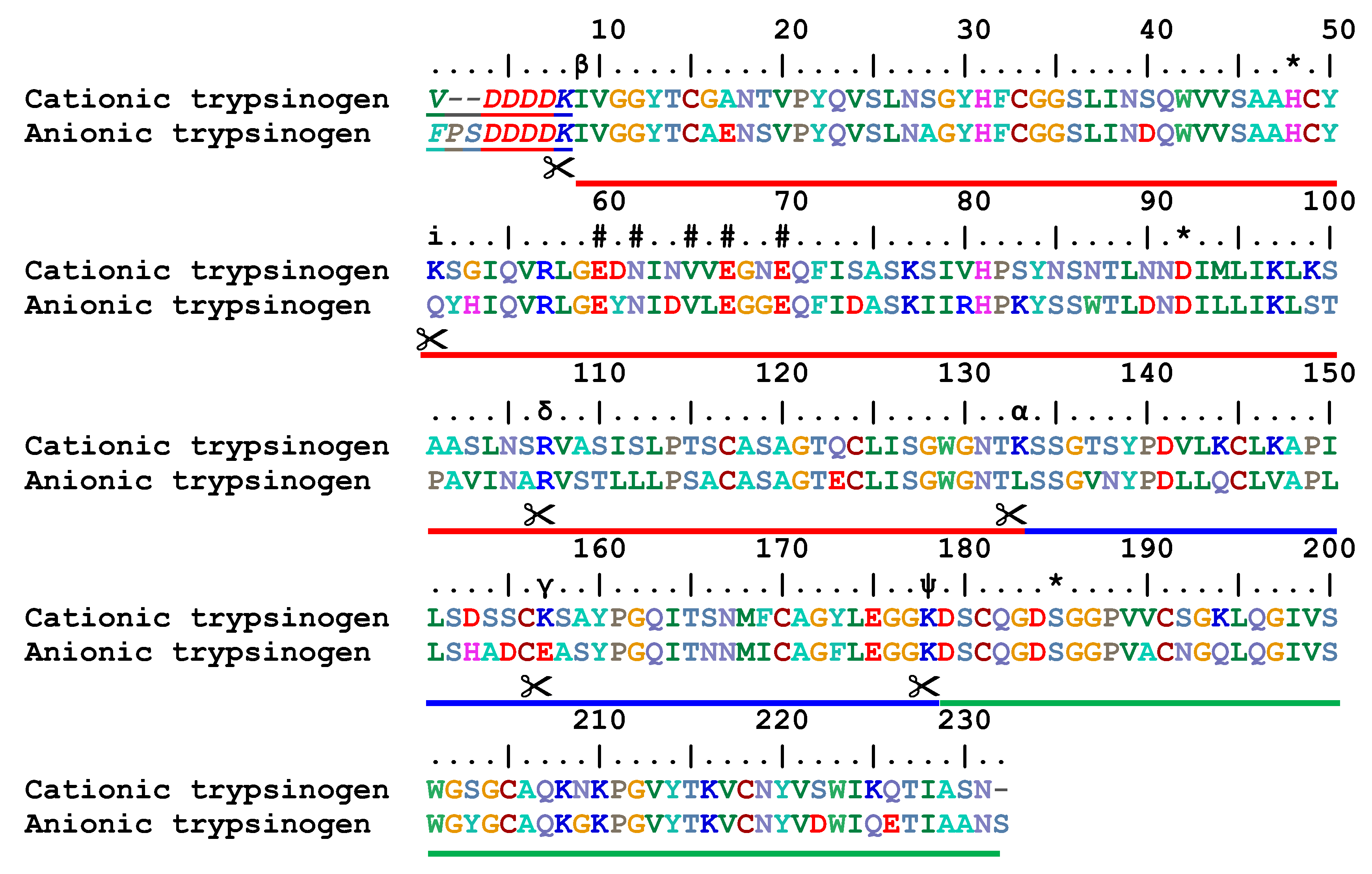
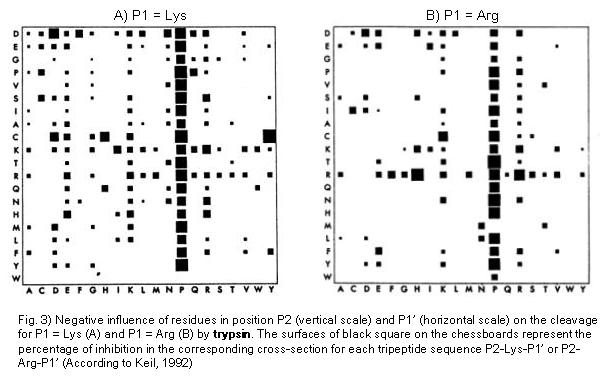
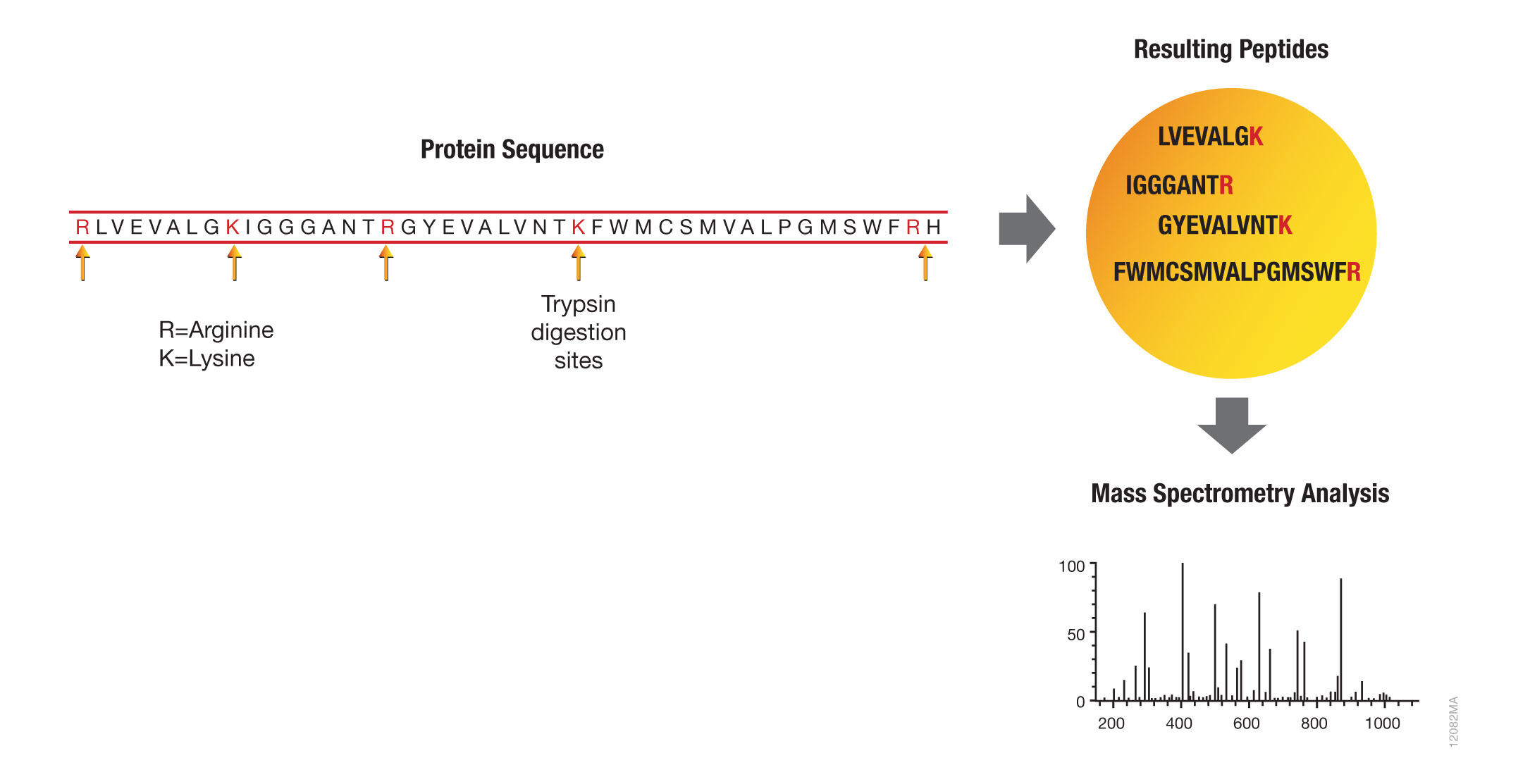
Schematic representation of the major trypsin cleavage sites and one... | Download Scientific Diagram
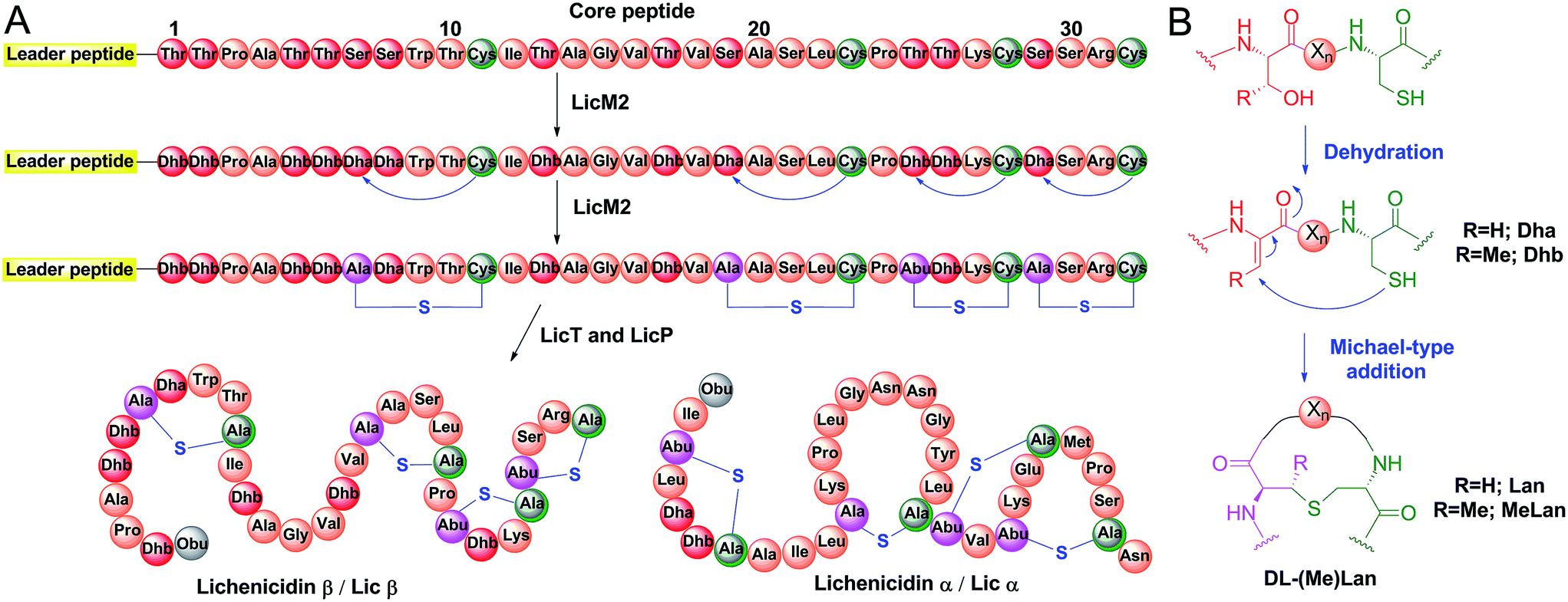
Applications of the class II lanthipeptide protease LicP for sequence-specific, traceless peptide bond cleavage - Chemical Science (RSC Publishing) DOI:10.1039/C5SC02329G

Protease cleavage site fingerprinting by label‐free in‐gel degradomics reveals pH‐dependent specificity switch of legumain | The EMBO Journal

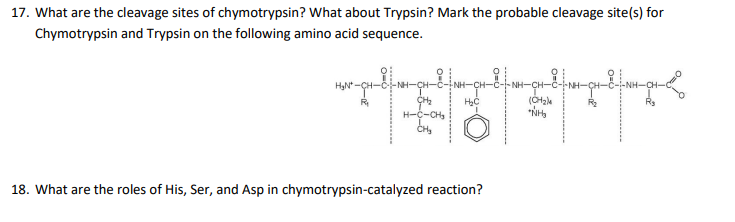
Marked difference in efficiency of the digestive enzymes pepsin, trypsin, chymotrypsin, and pancreatic elastase to cleave tightly folded proteins
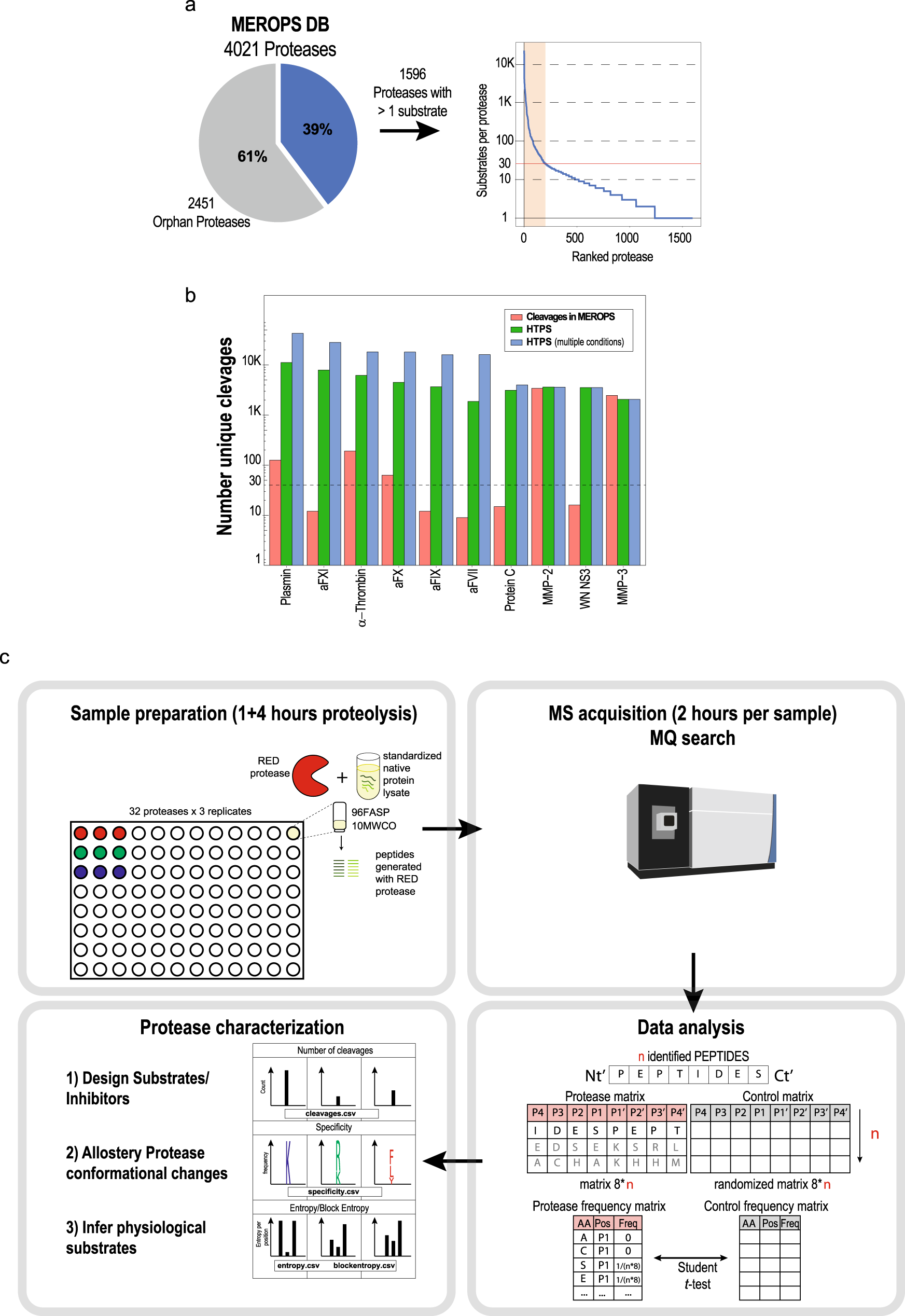
Mapping specificity, cleavage entropy, allosteric changes and substrates of blood proteases in a high-throughput screen | Nature Communications

Marked difference in efficiency of the digestive enzymes pepsin, trypsin, chymotrypsin, and pancreatic elastase to cleave tightly folded proteins

Proteomic Identification of Protease Cleavage Sites Characterizes Prime and Non-prime Specificity of Cysteine Cathepsins B, L, and S | Journal of Proteome Research

Global Sequencing of Proteolytic Cleavage Sites in Apoptosis by Specific Labeling of Protein N Termini: Cell