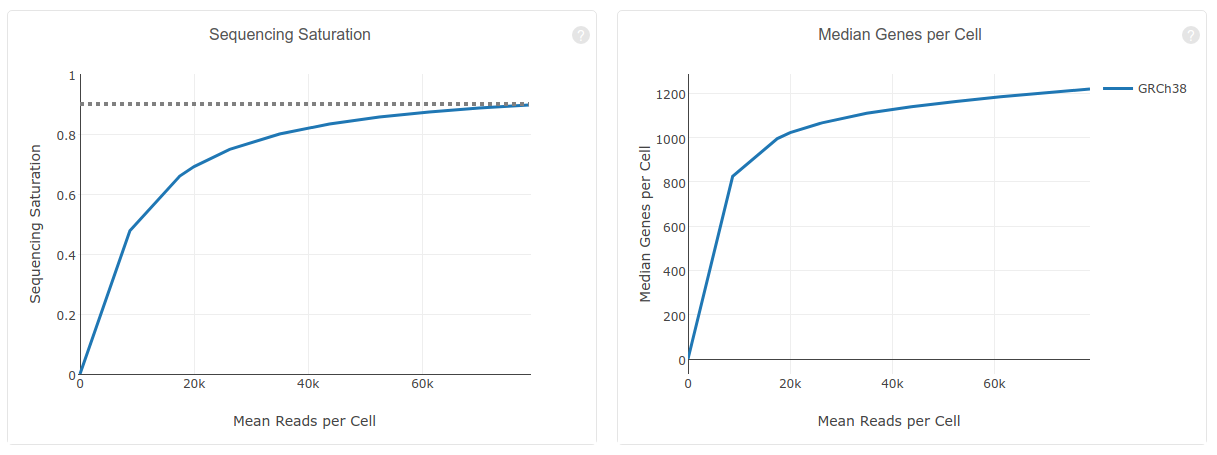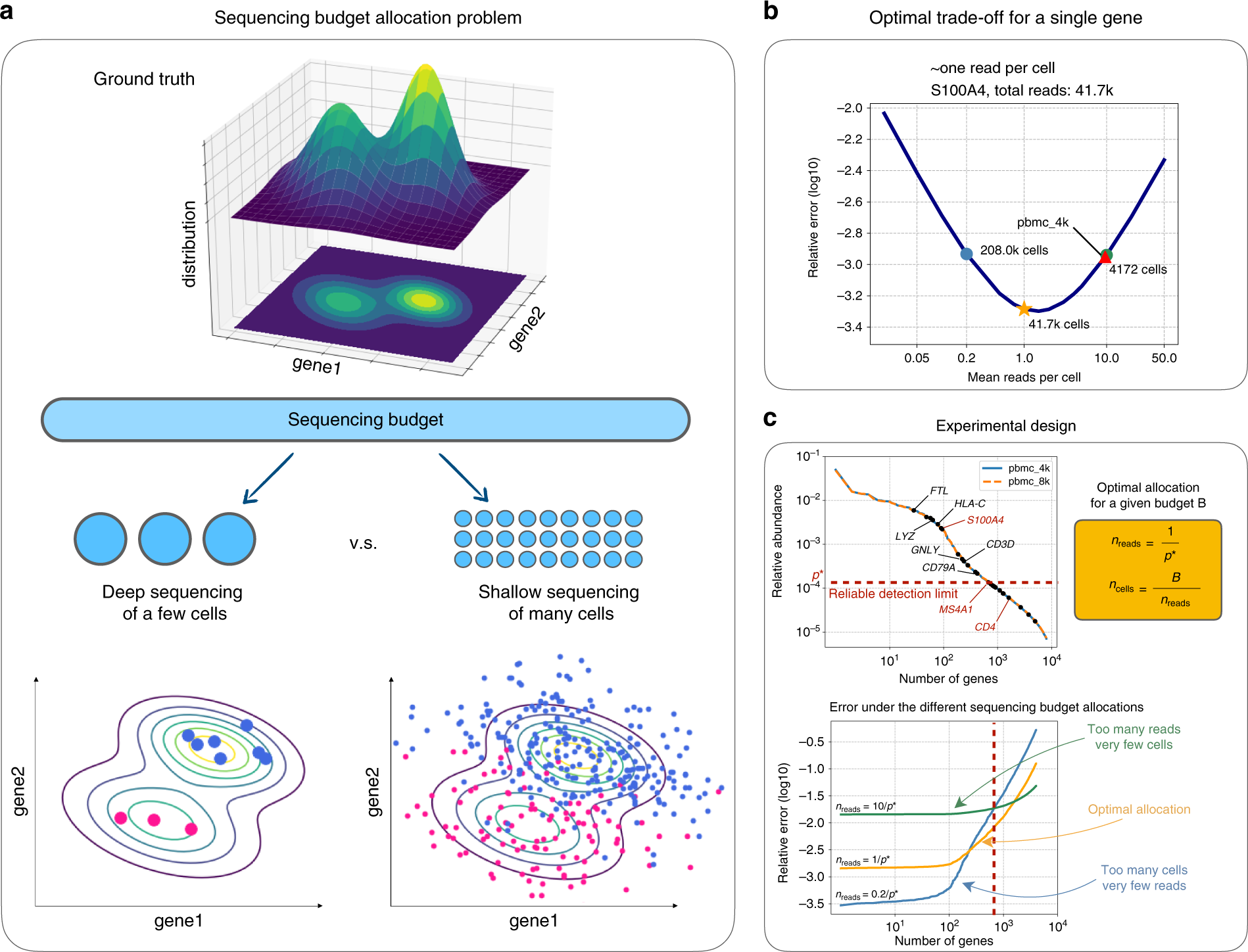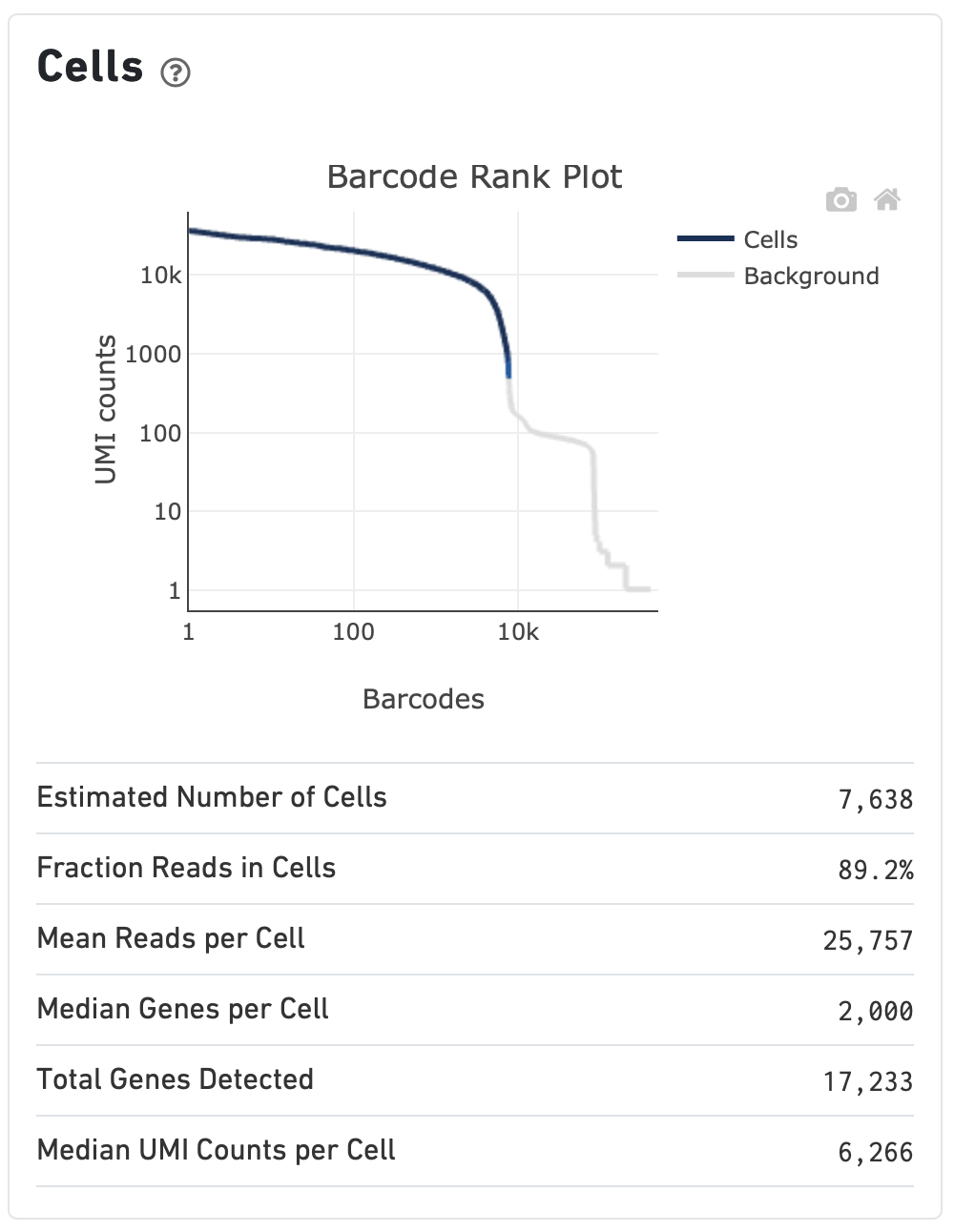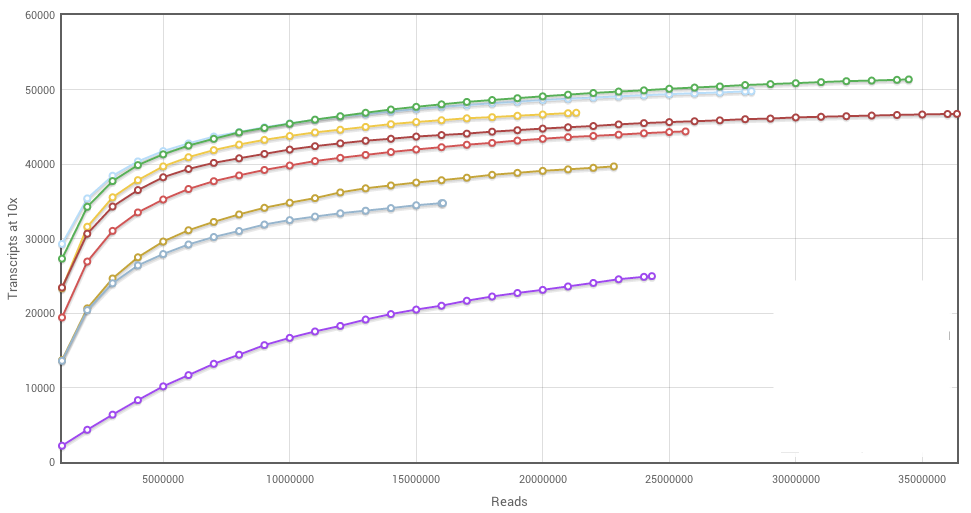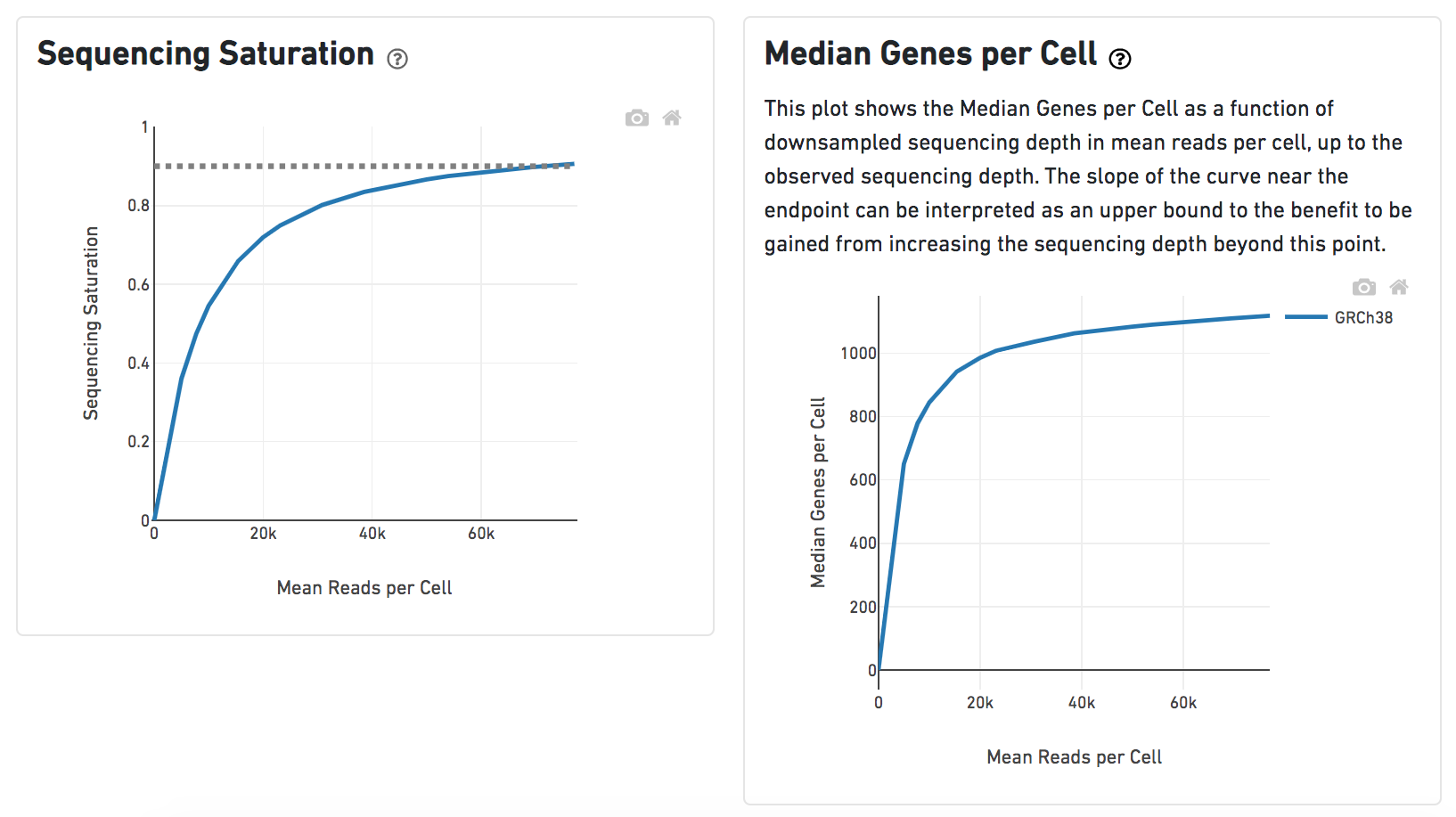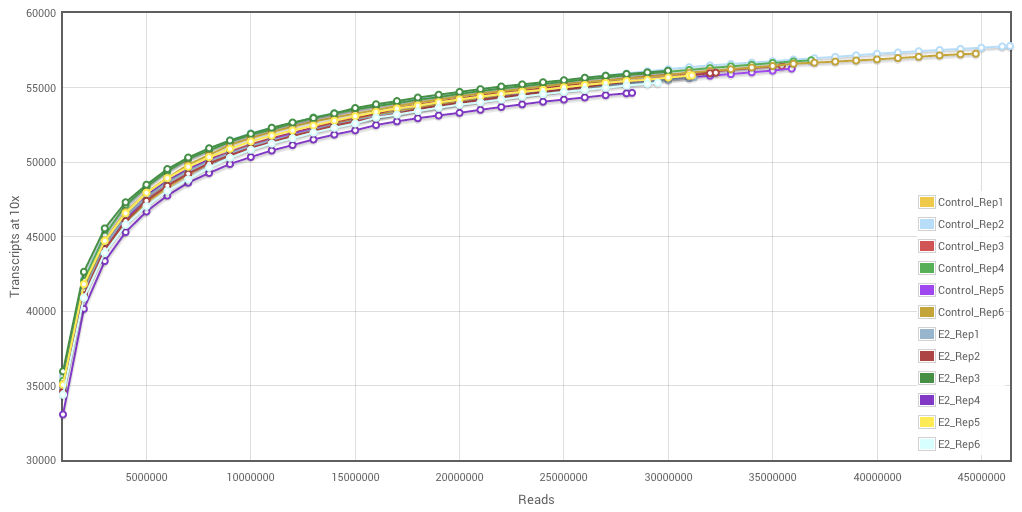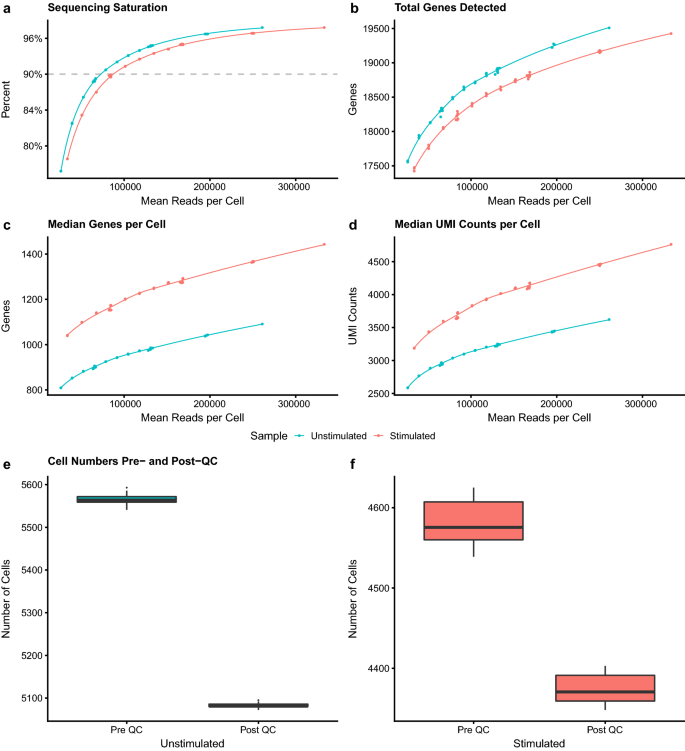
Characterisation of CD4+ T-cell subtypes using single cell RNA sequencing and the impact of cell number and sequencing depth | Scientific Reports

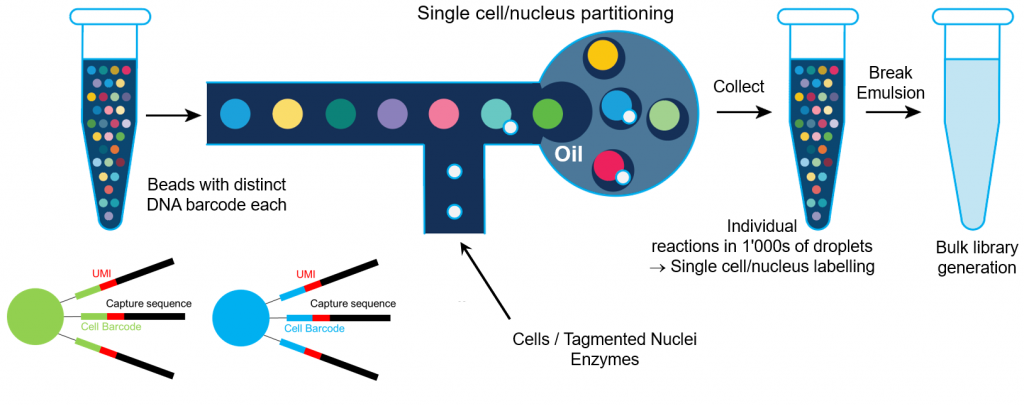
Simultaneous trimodal single-cell measurement of transcripts, epitopes, and chromatin accessibility using TEA-seq | eLife

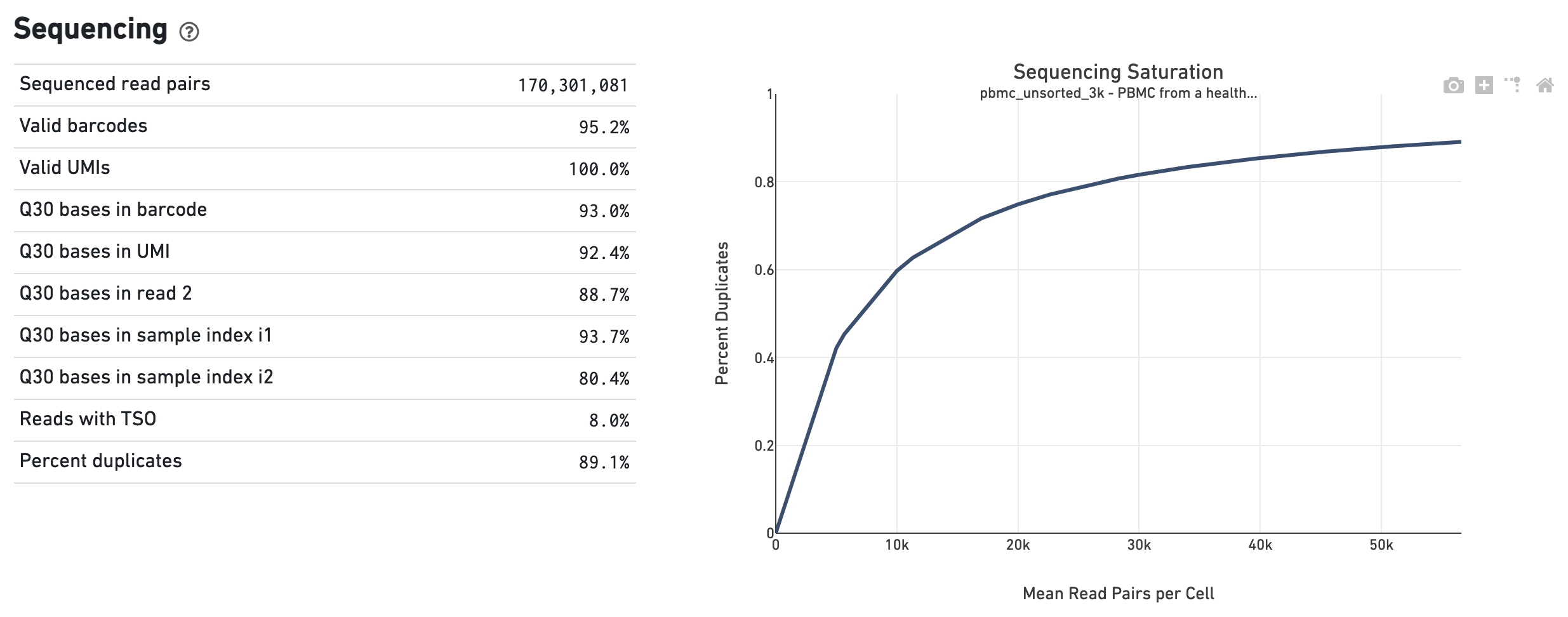
Experimental Design for Immune Profiling -Software -Single Cell Immune Profiling -Official 10x Genomics Support

HyPR-seq: Single-cell quantification of chosen RNAs via hybridization and sequencing of DNA probes | PNAS