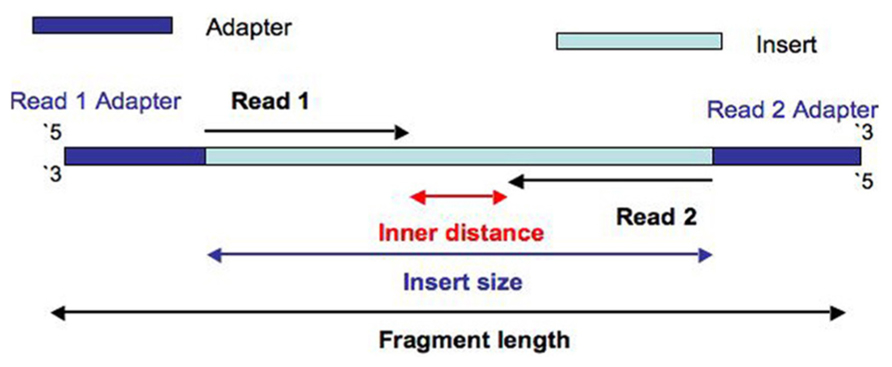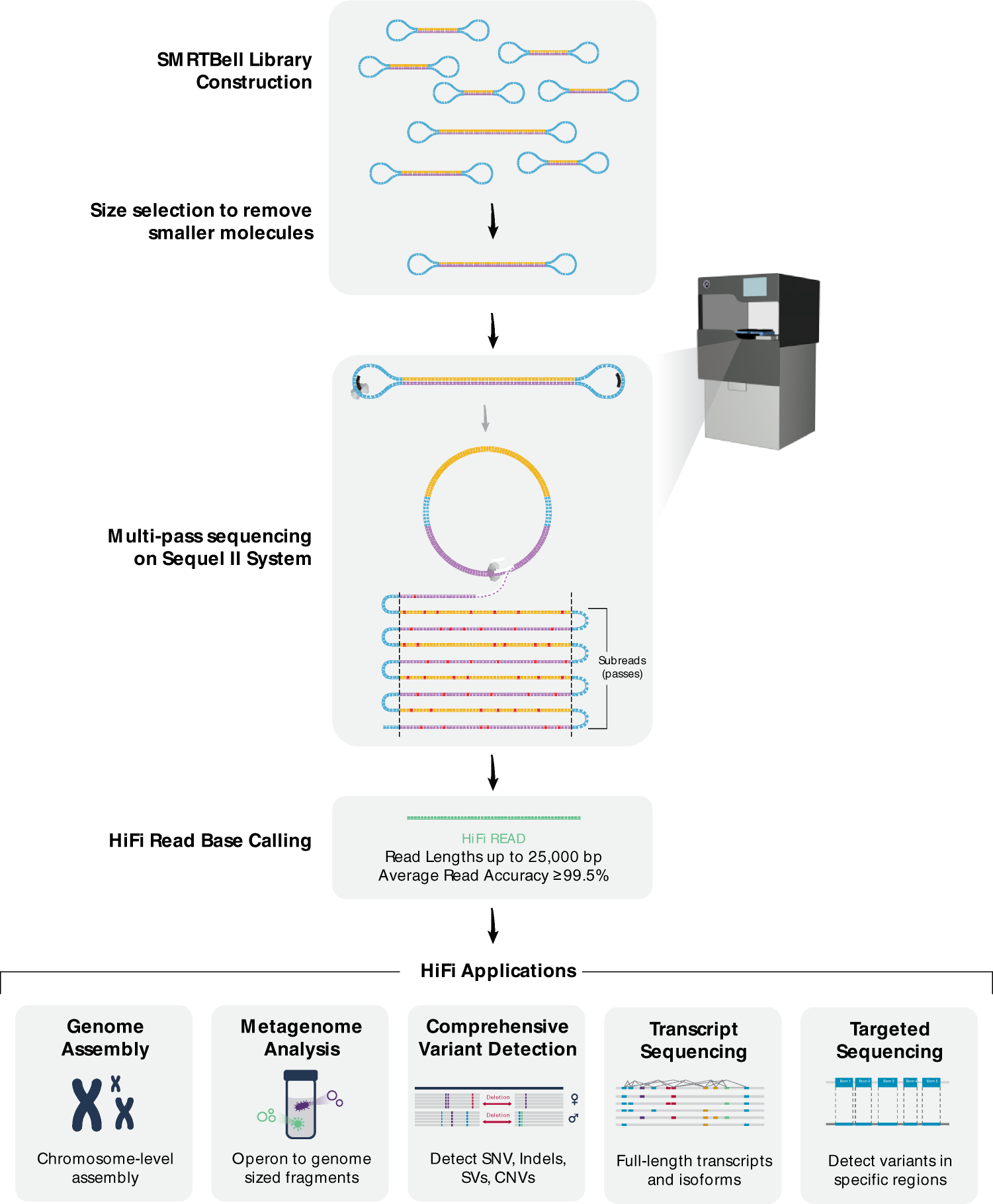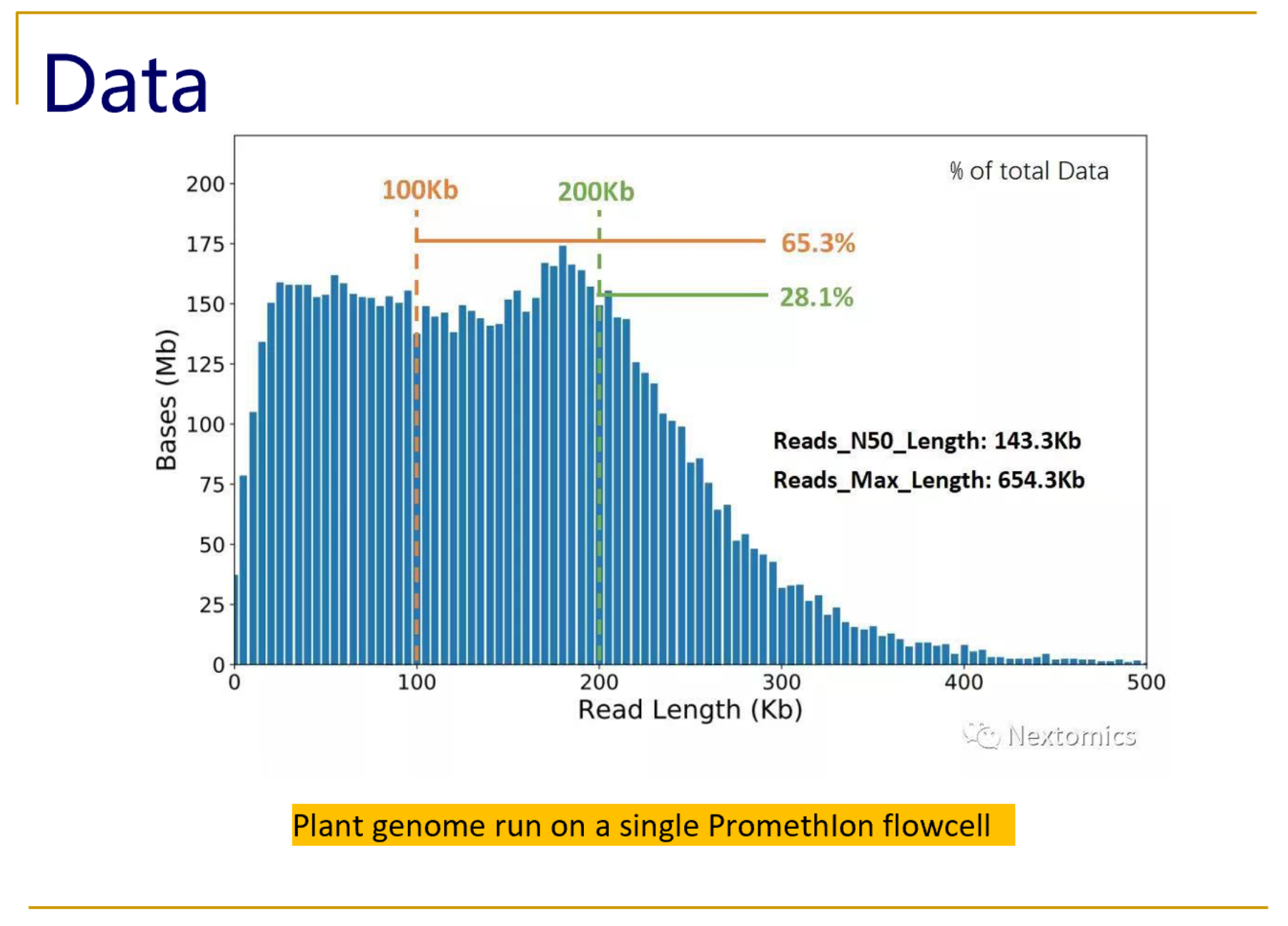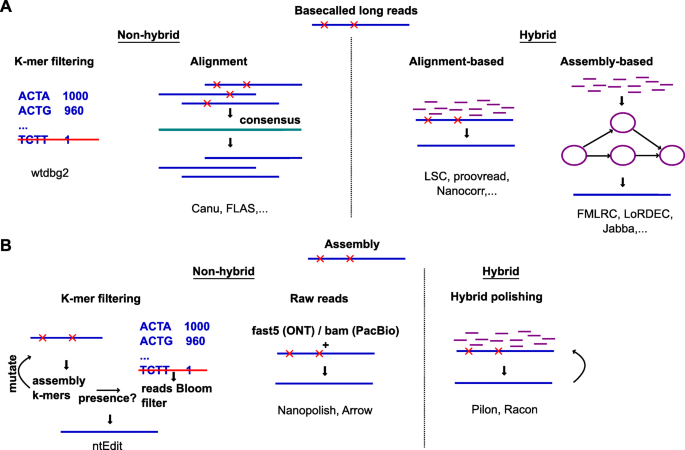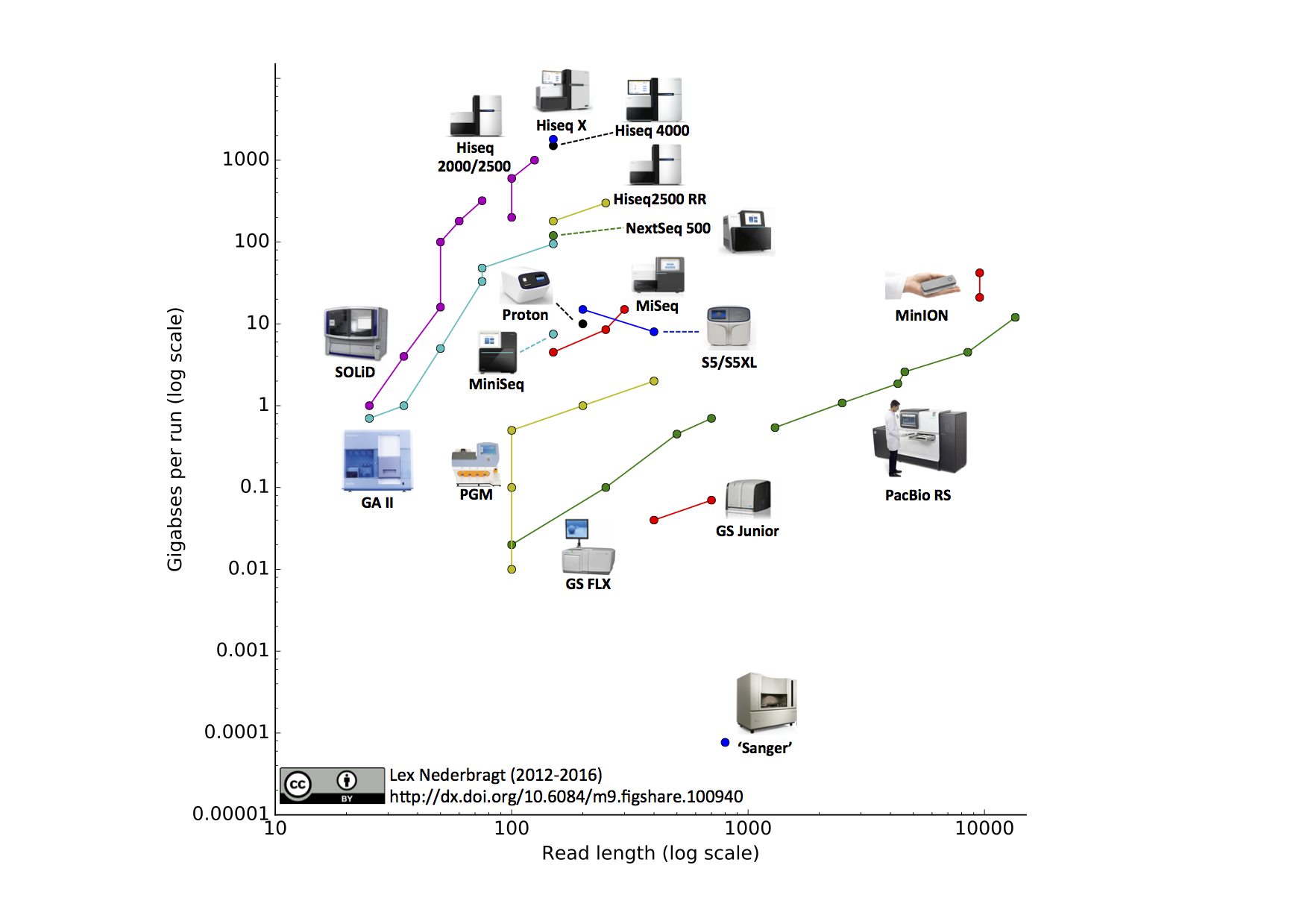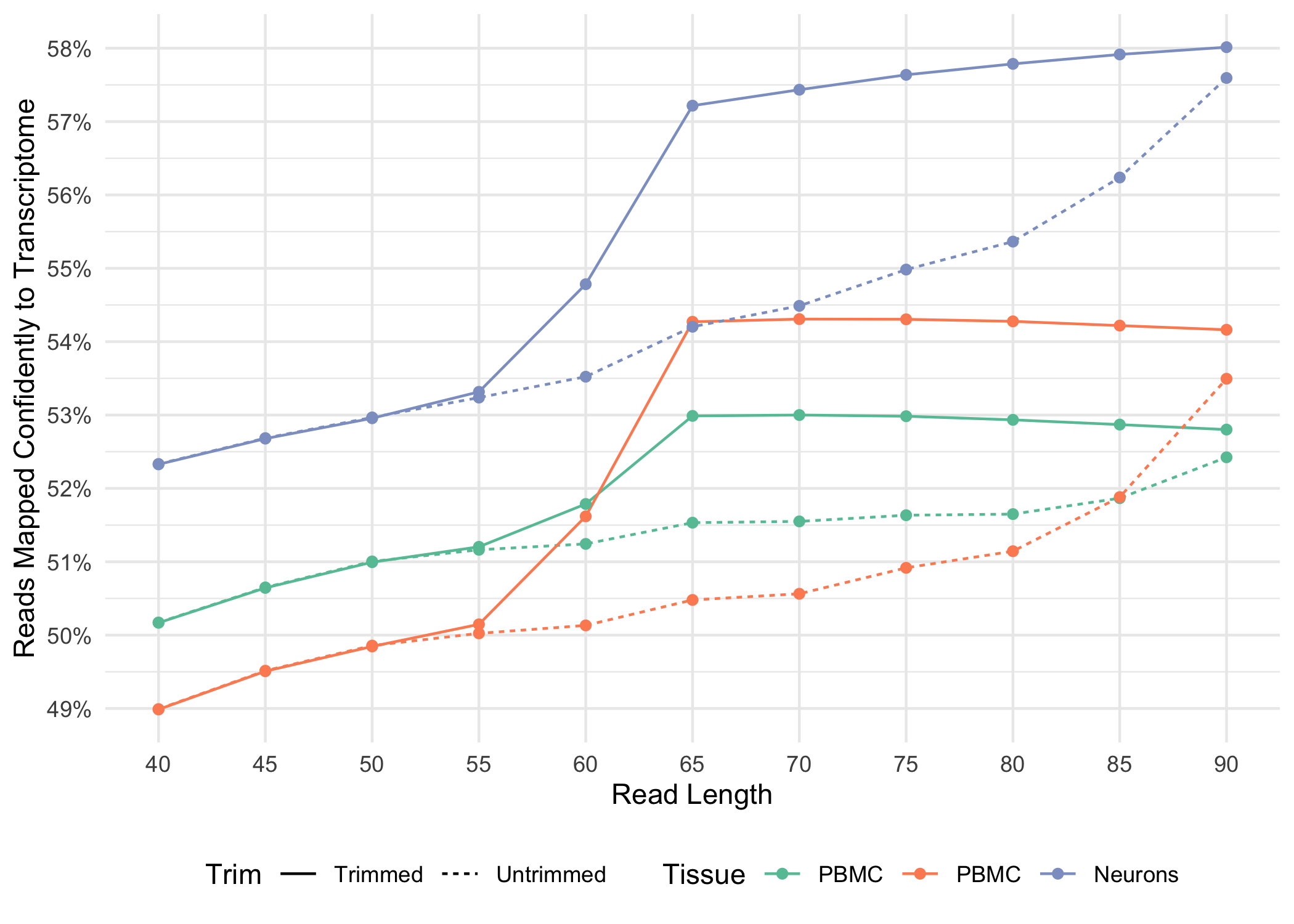
Can I decrease the sequencing length of Read 2 for Single Cell Gene Expression assays? – 10X Genomics
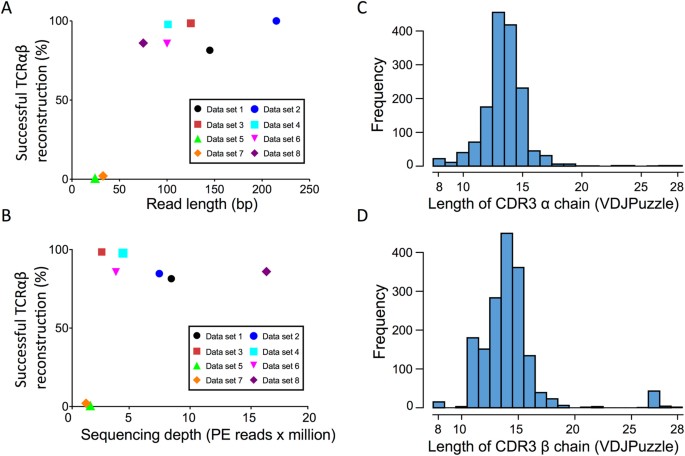
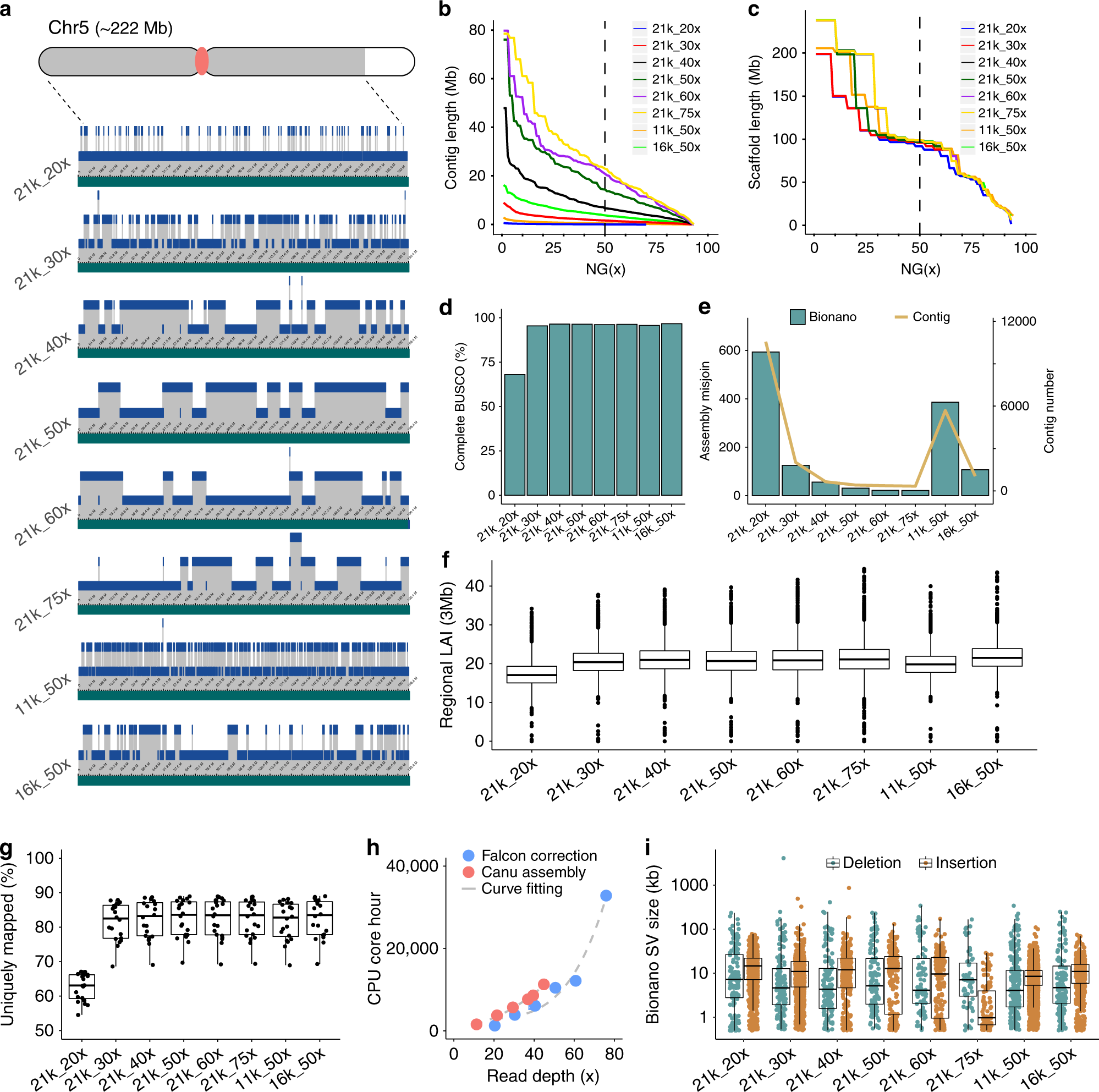
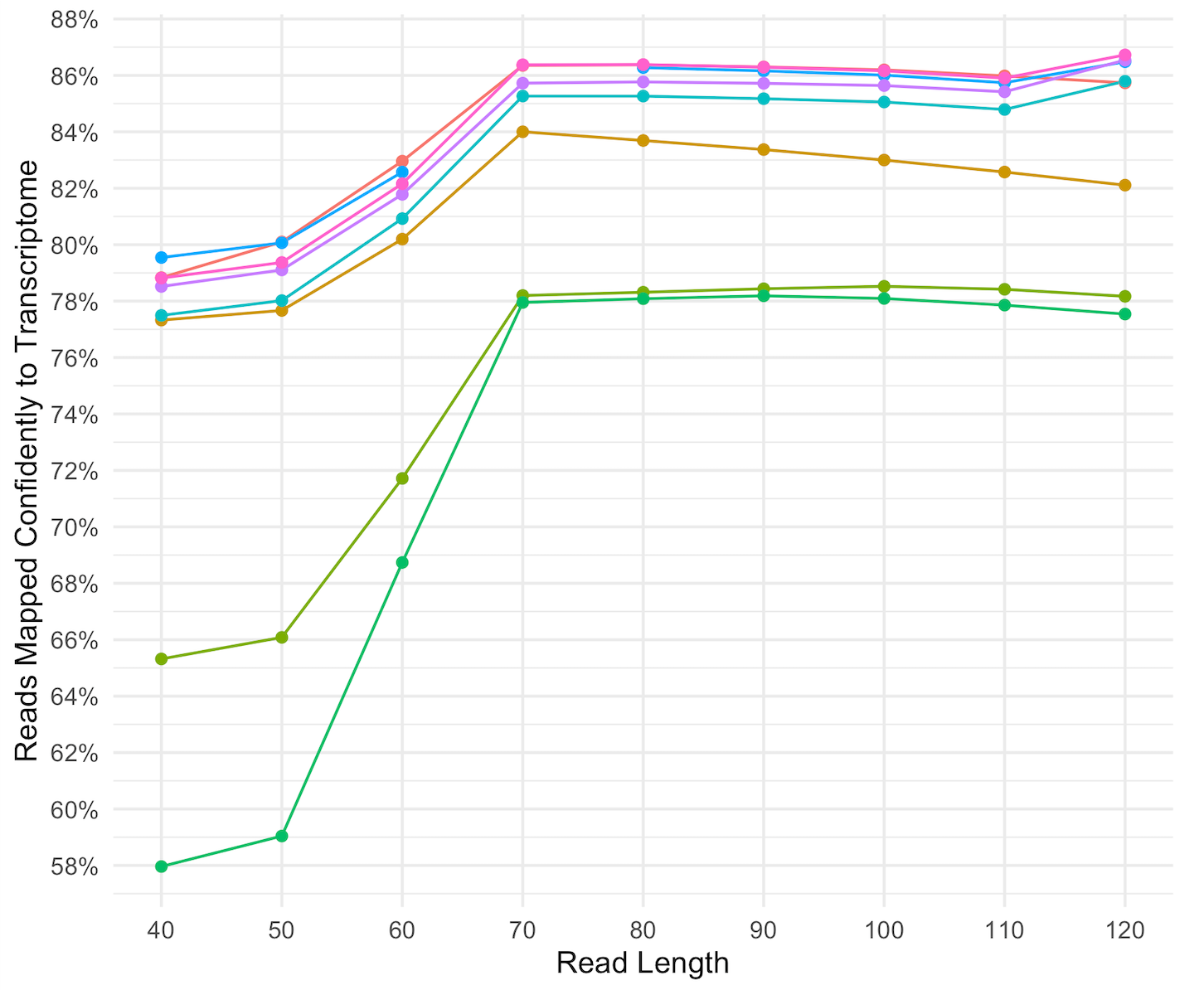
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports

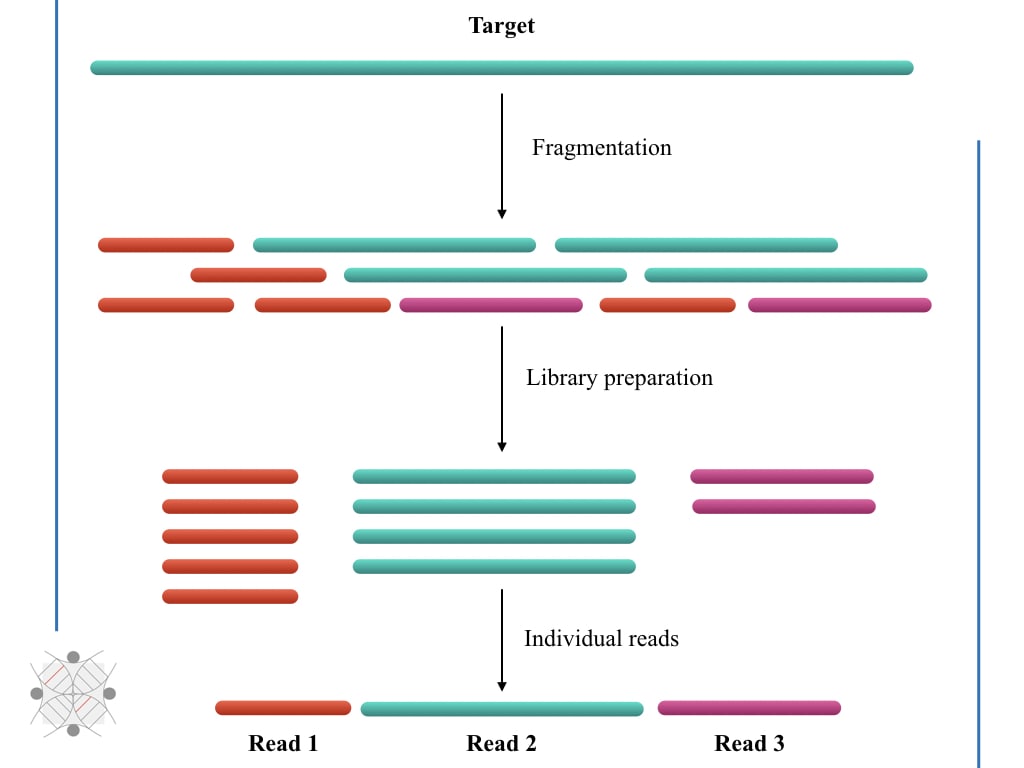
Oxford Nanopore Technology: A Promising Long-Read Sequencing Platform To Study Exon Connectivity and Characterize Isoforms of Complex Genes | Semantic Scholar