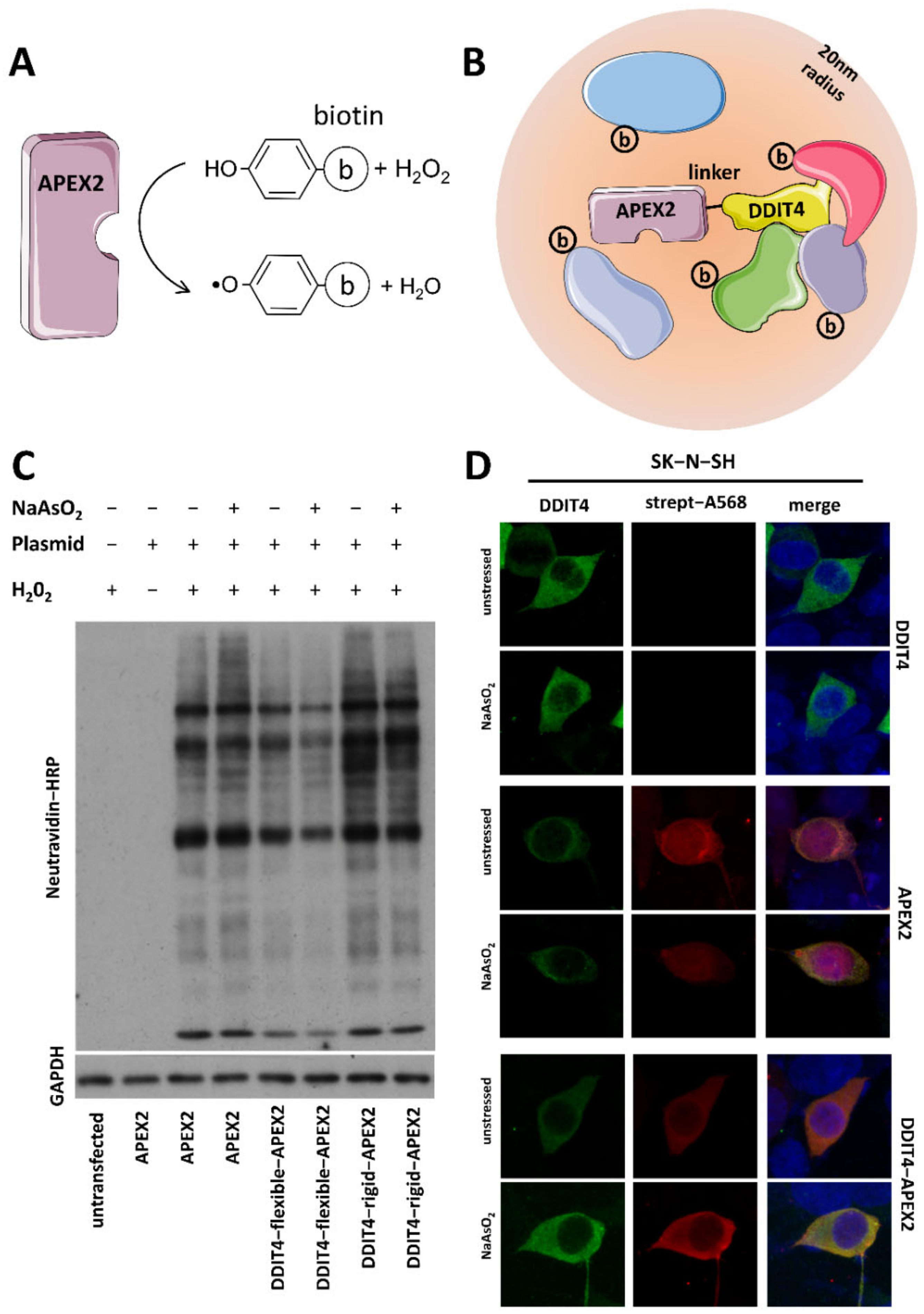
SBF-SEM and FIB-SEM analysis of Erg11-APEX2 in S. cerevisiae. (A, B,... | Download Scientific Diagram

Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules - ScienceDirect

Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules - ScienceDirect
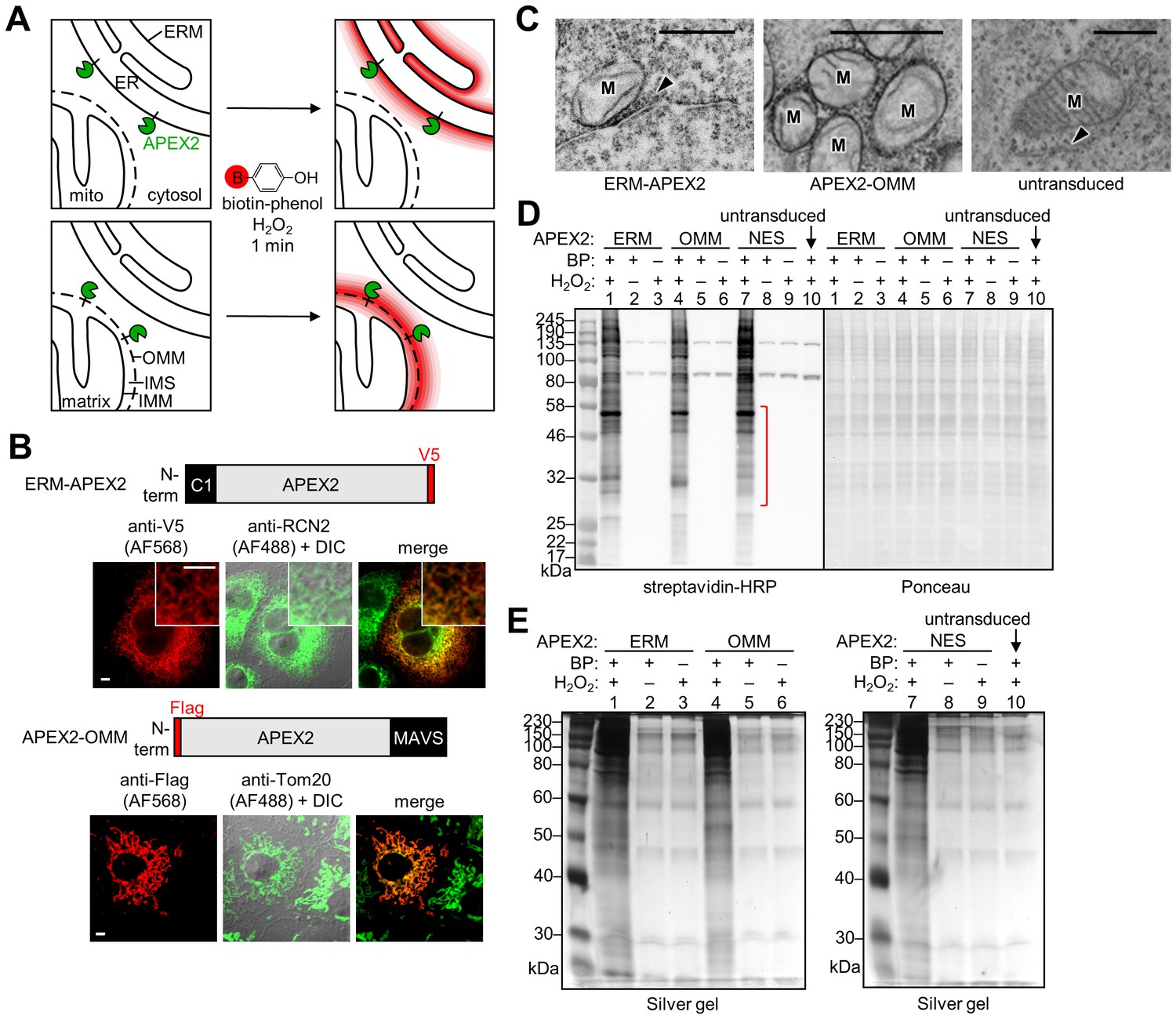
Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation | eLife
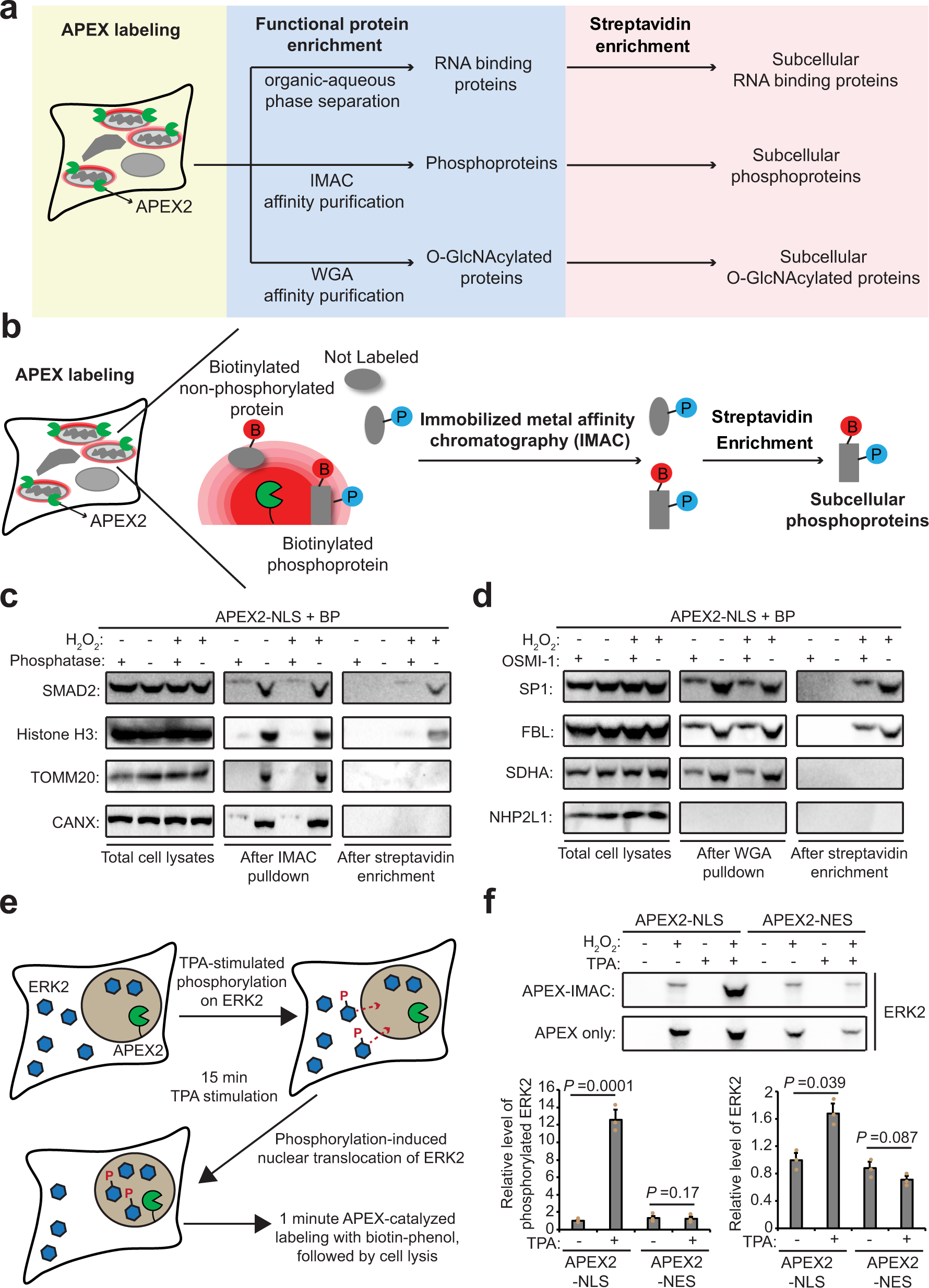
Spatiotemporally-resolved mapping of RNA binding proteins via functional proximity labeling reveals a mitochondrial mRNA anchor promoting stress recovery | Nature Communications
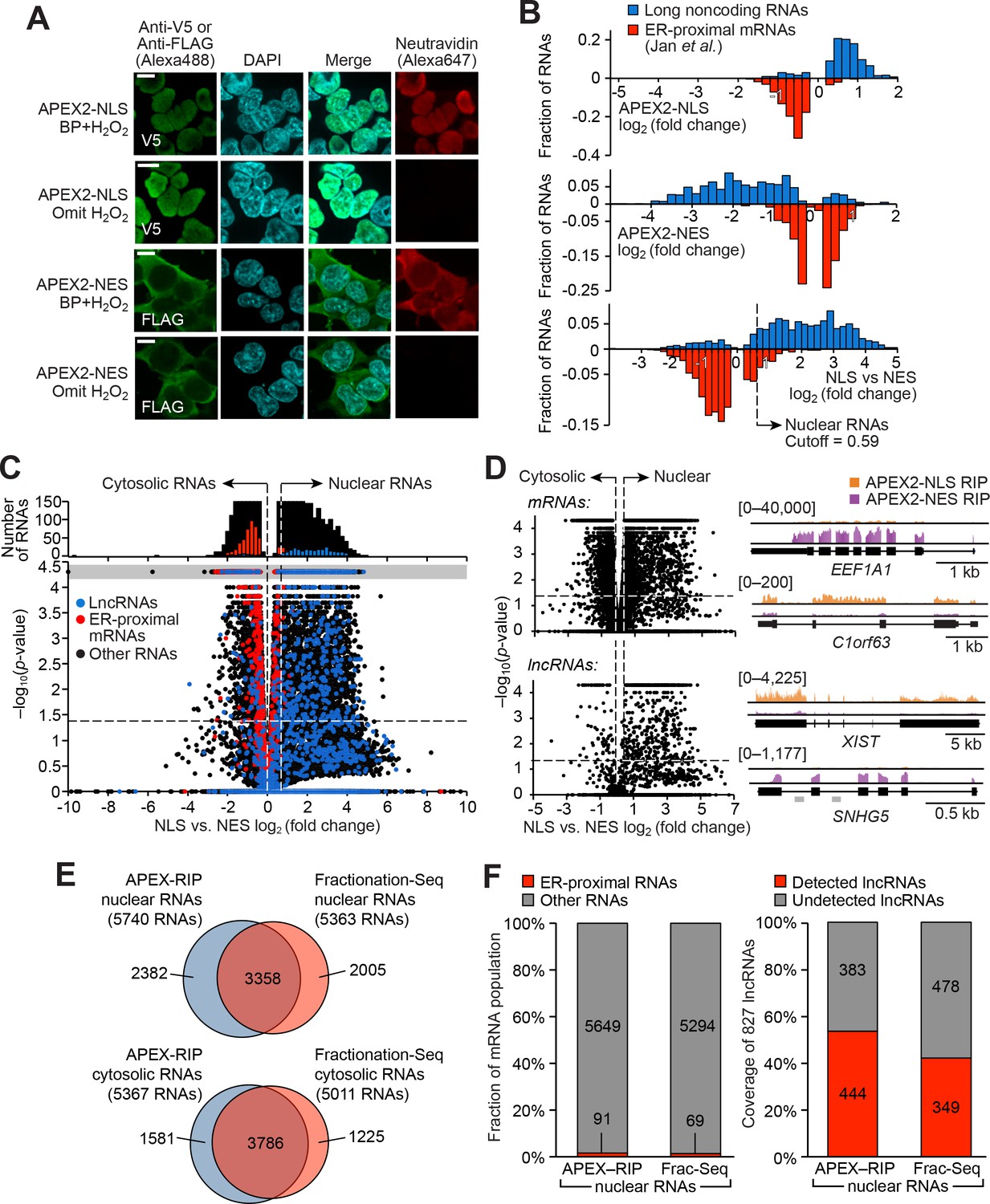
Figures and data in Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking | eLife
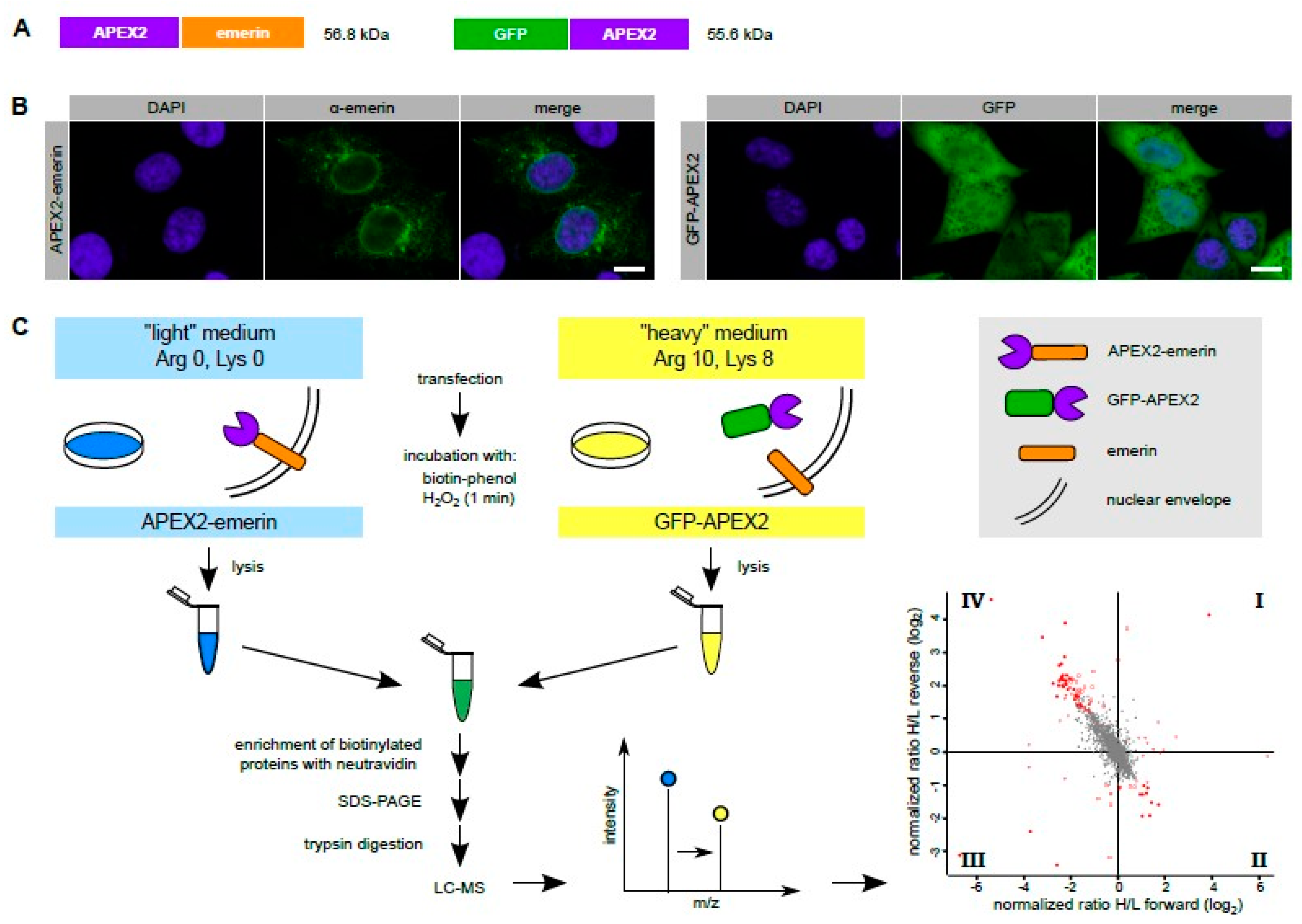
Cells | Free Full-Text | Probing the Environment of Emerin by Enhanced Ascorbate Peroxidase 2 (APEX2)-Mediated Proximity Labeling

APEX2‐mediated RAB proximity labeling identifies a role for RAB21 in clathrin‐independent cargo sorting | EMBO reports