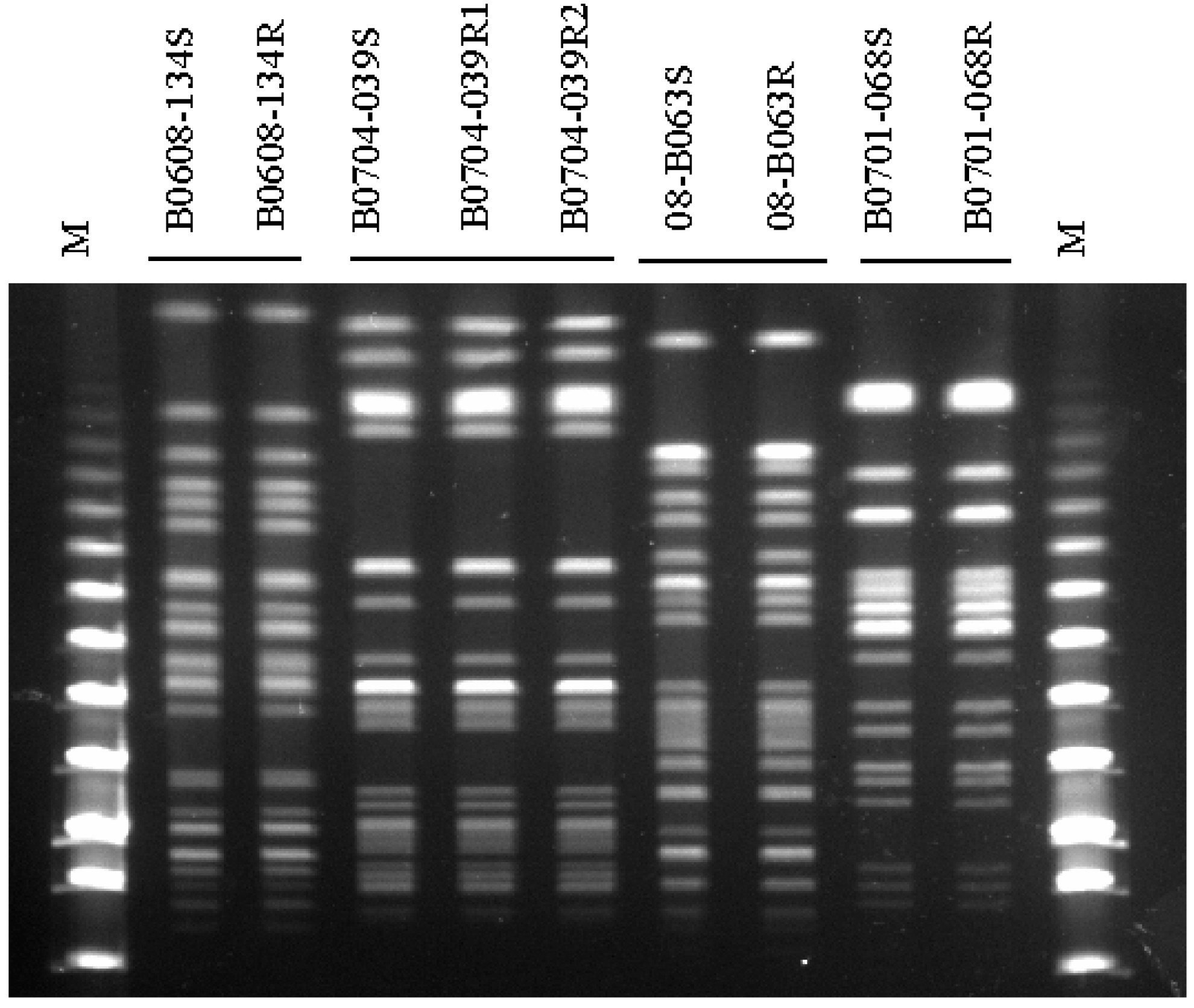
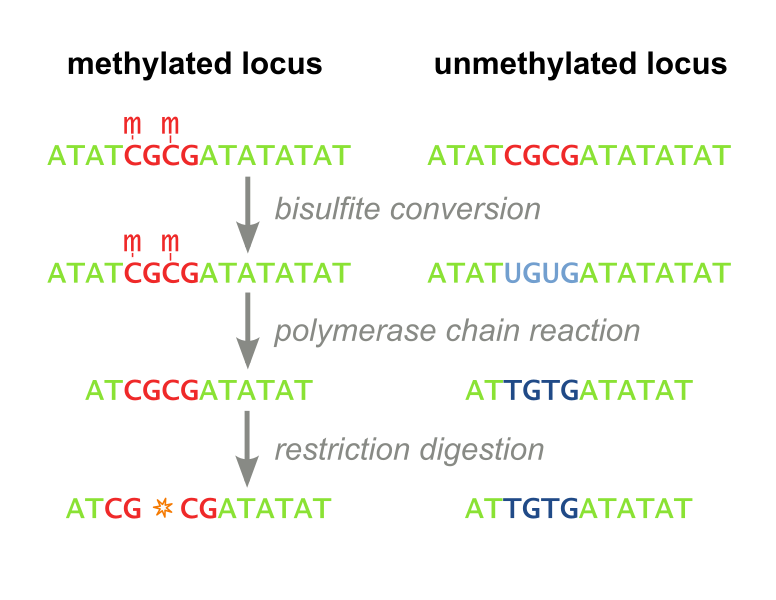
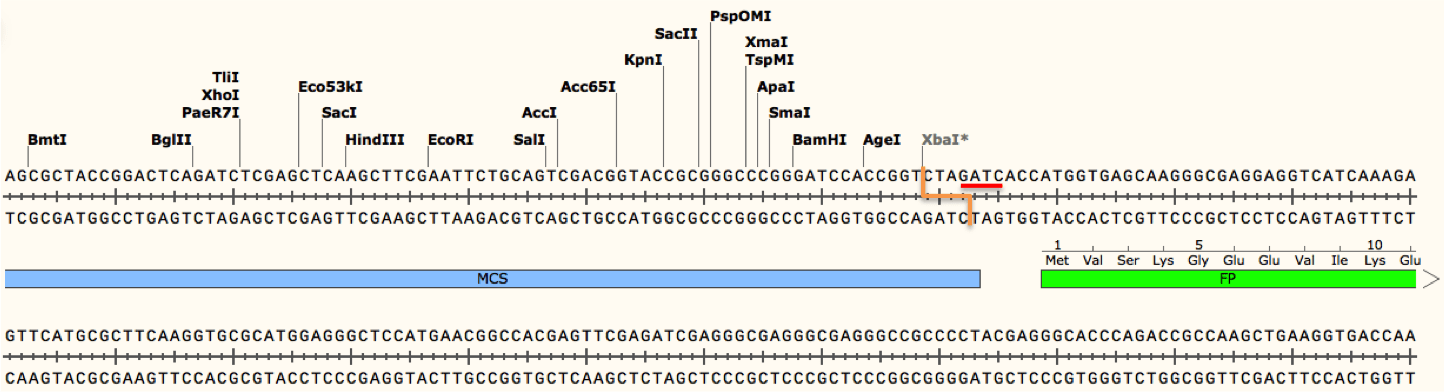
In Vitro Seamless Stack Enzymatic Assembly of DNA Molecules Based on a Strategy Involving Splicing of Restriction Sites | Scientific Reports
DNA sequences of the crossover regions of EcoRI/XbaI-digested pUC19 and... | Download Scientific Diagram
3 Multiple cloning site of pMluI vector. EcoRI and XbaI, restriction... | Download Scientific Diagram
![PDF] Quick identification of Type I restriction enzyme isoschizomers using newly developed pTypeI and reference plasmids | Semantic Scholar PDF] Quick identification of Type I restriction enzyme isoschizomers using newly developed pTypeI and reference plasmids | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/0c28e2307444232d8005a49ff7d76c63db1fa2bb/3-Figure1-1.png)
PDF] Quick identification of Type I restriction enzyme isoschizomers using newly developed pTypeI and reference plasmids | Semantic Scholar
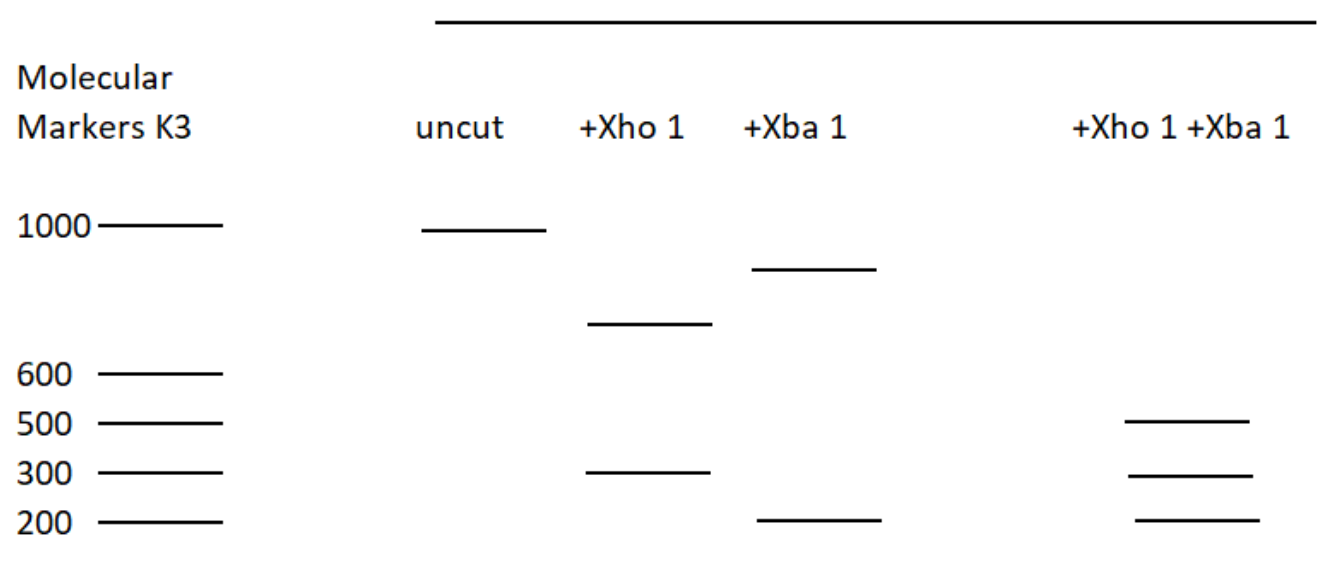
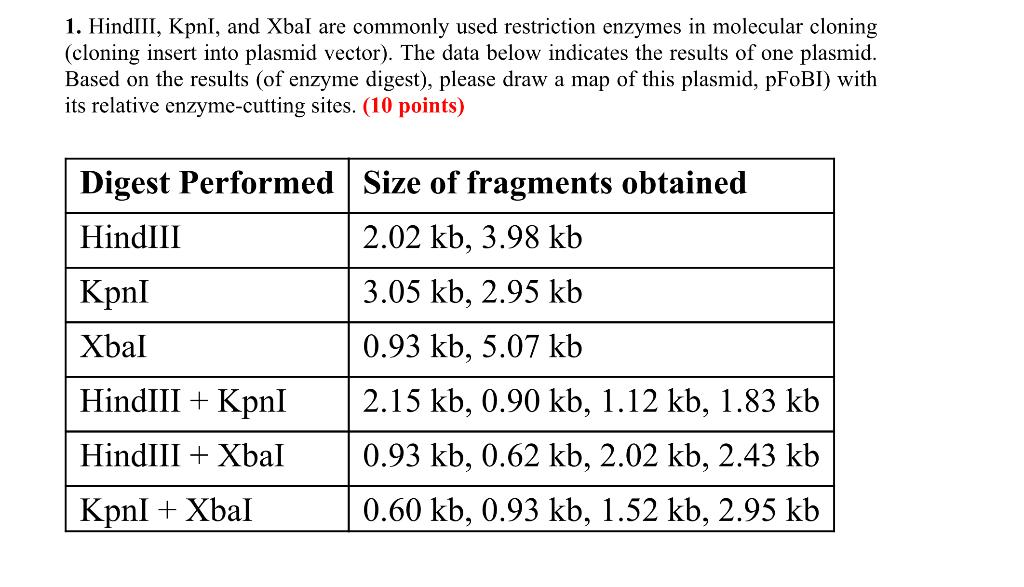
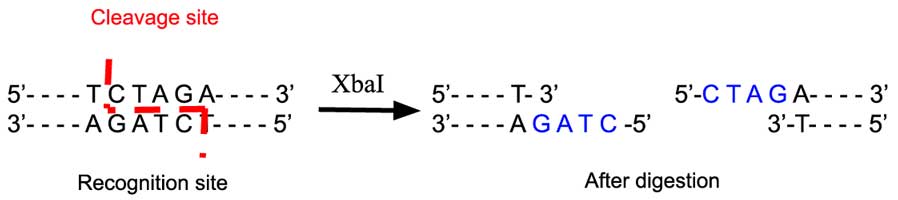
The restriction enzyme XhoI and XbaI were used to digest a linear fragment of DNA at a specific sequence such as that shown for EcoR1 (figure above). Use the information gained from