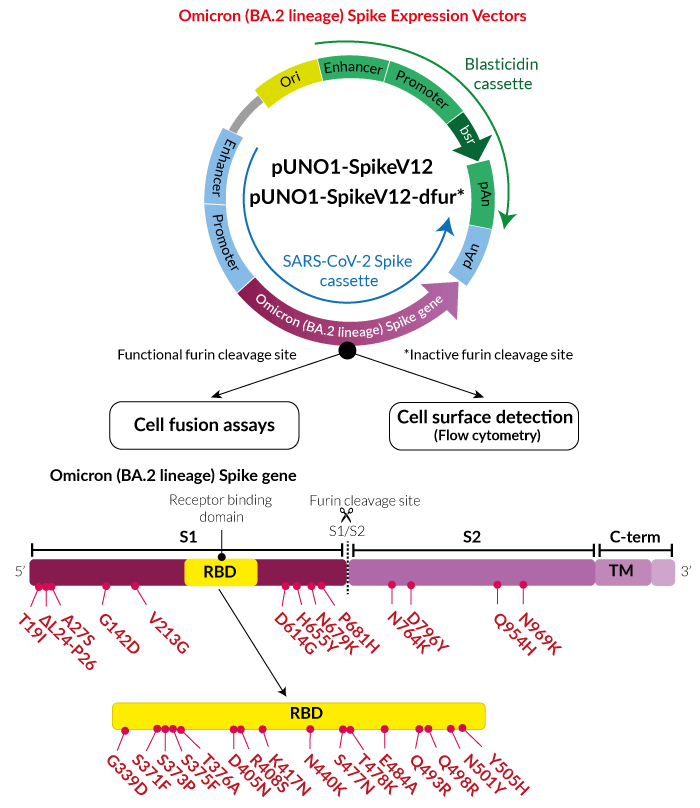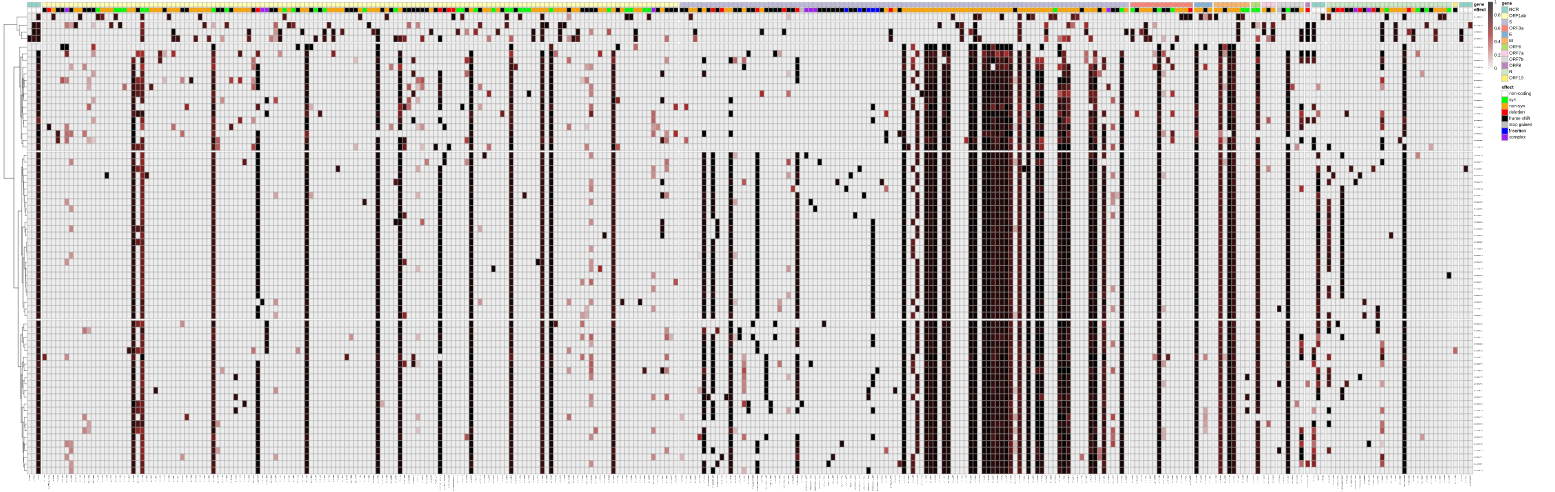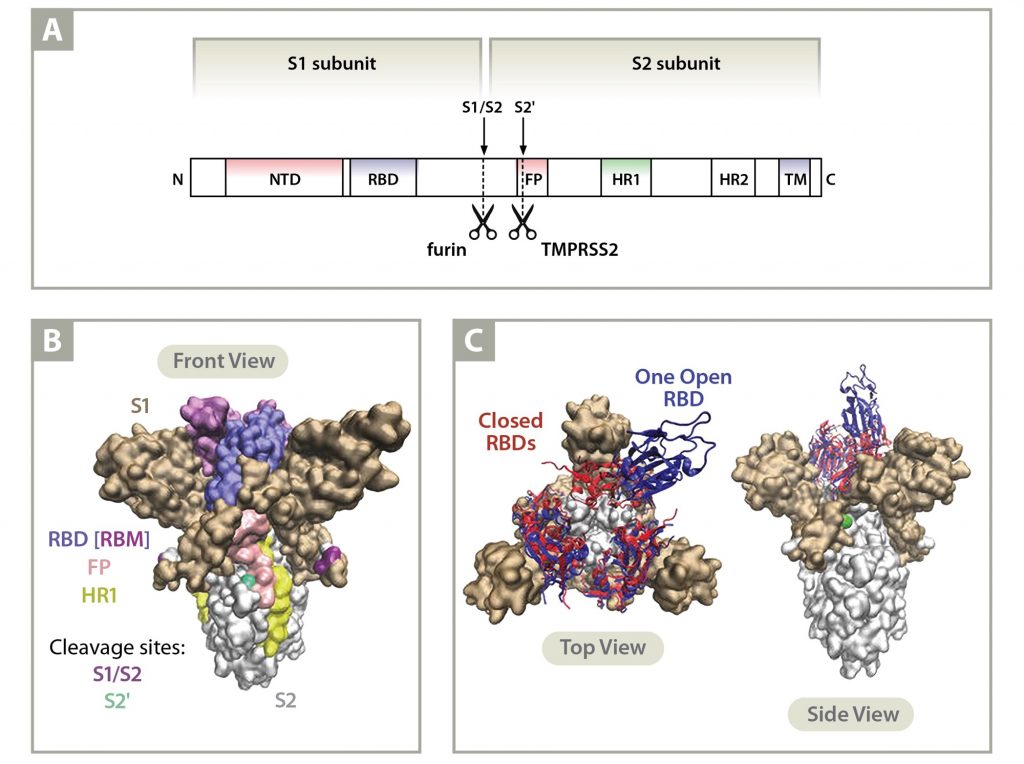
Implications of the Mutations in the Spike Protein of the Omicron Variant of Concern (VoC) of SARS-CoV-2 – Signature Science

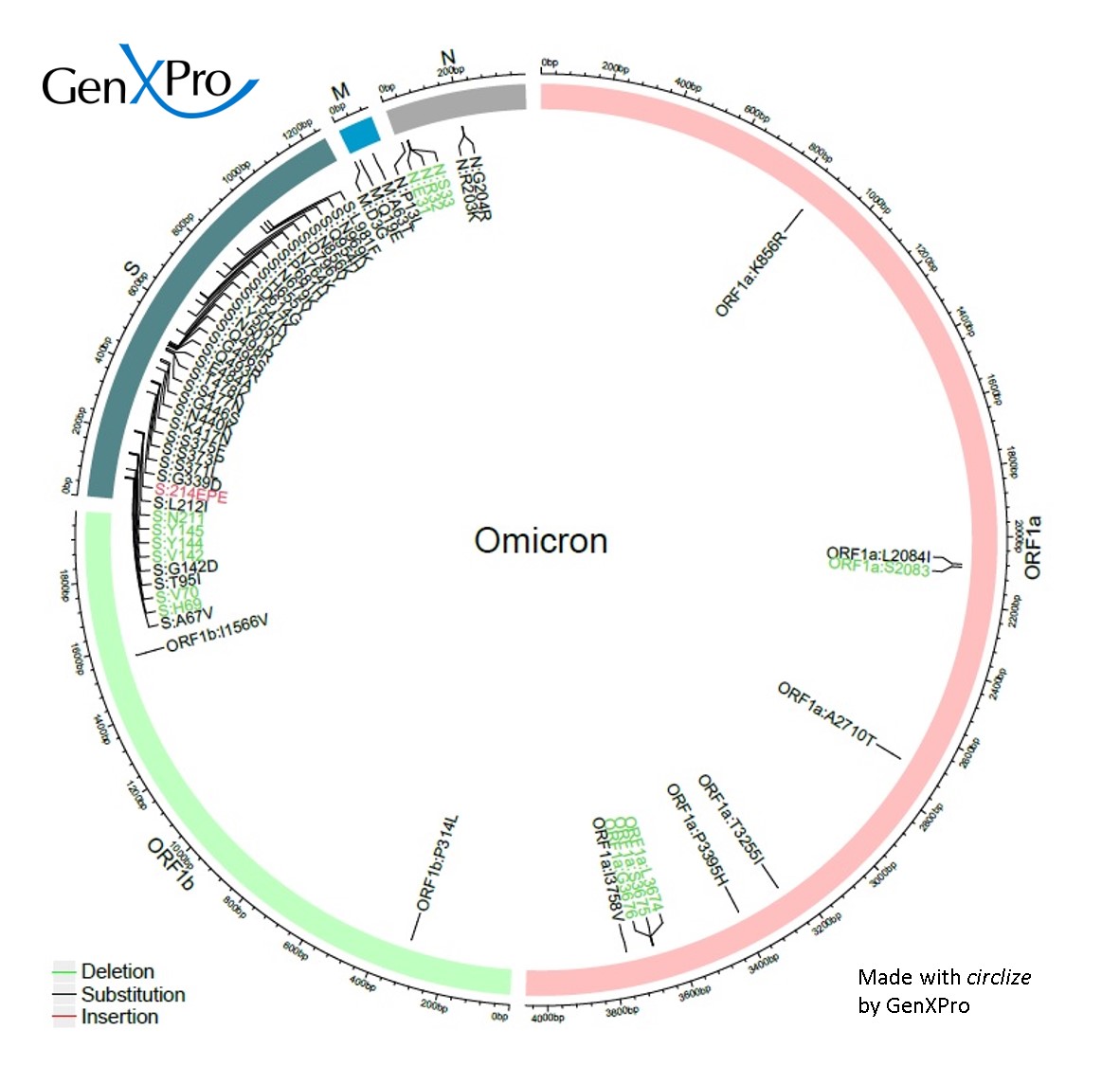
Sequence analysis of the emerging SARS‐CoV‐2 variant Omicron in South Africa - Wang - 2022 - Journal of Medical Virology - Wiley Online Library
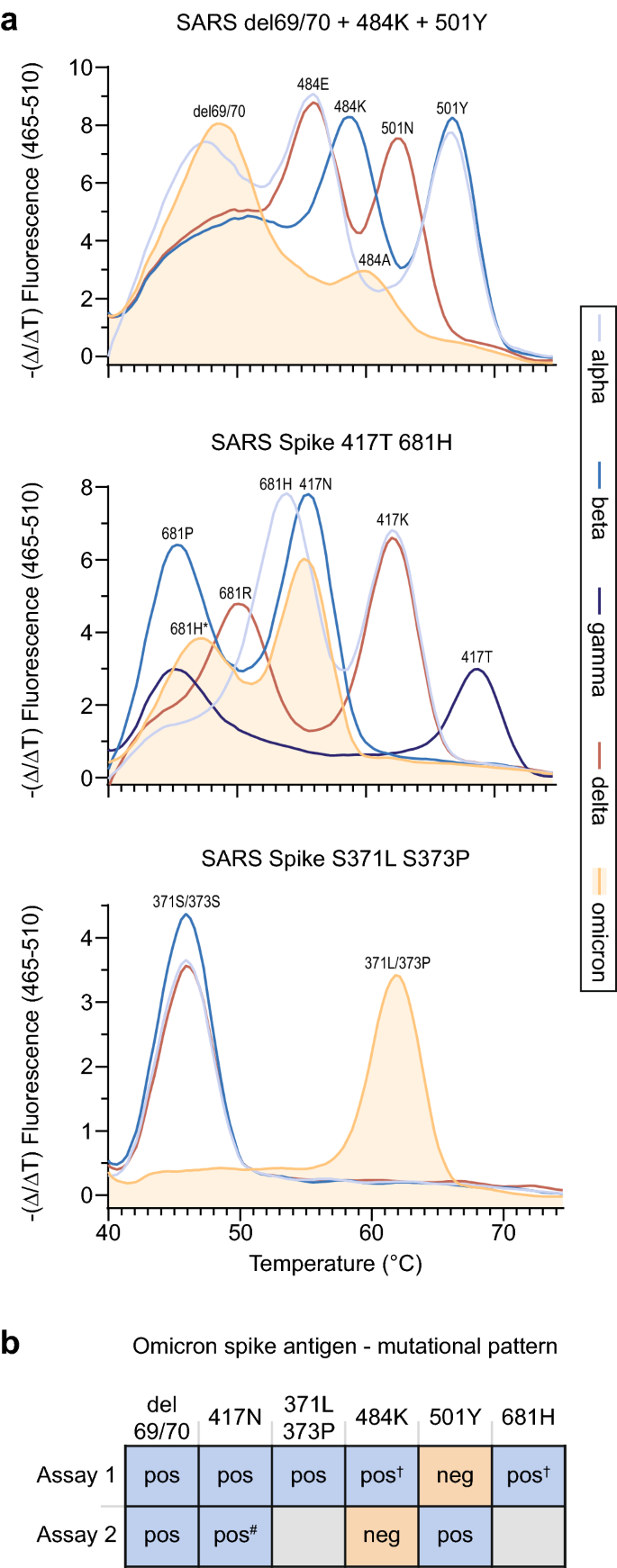
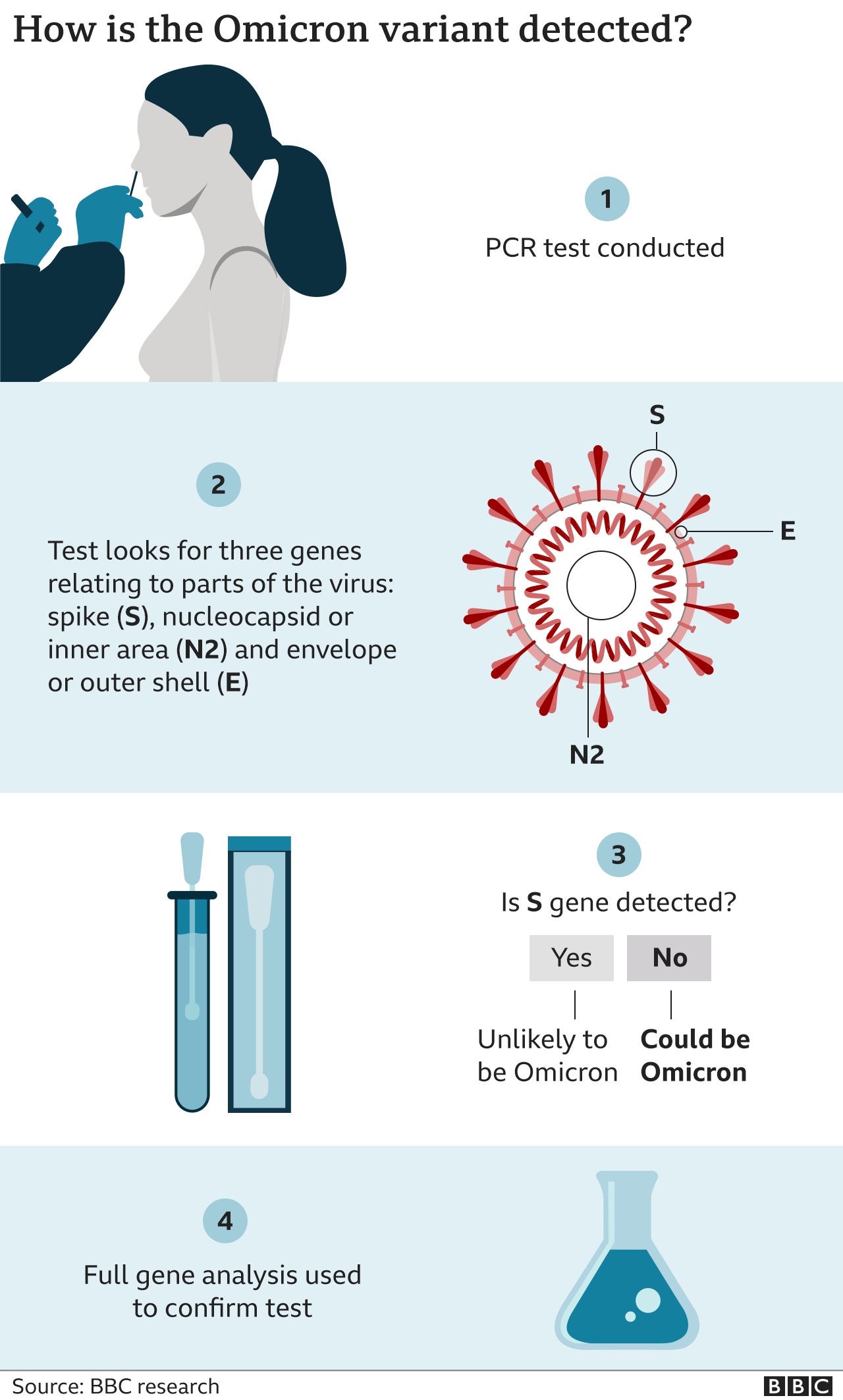
Rapid and sensitive identification of omicron by variant-specific PCR and nanopore sequencing: paradigm for diagnostics of emerging SARS-CoV-2 variants | SpringerLink

Genes | Free Full-Text | SARS-CoV-2 Omicron Variant Genomic and Phylogenetic Analysis in Iraqi Kurdistan Region

SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses - ScienceDirect
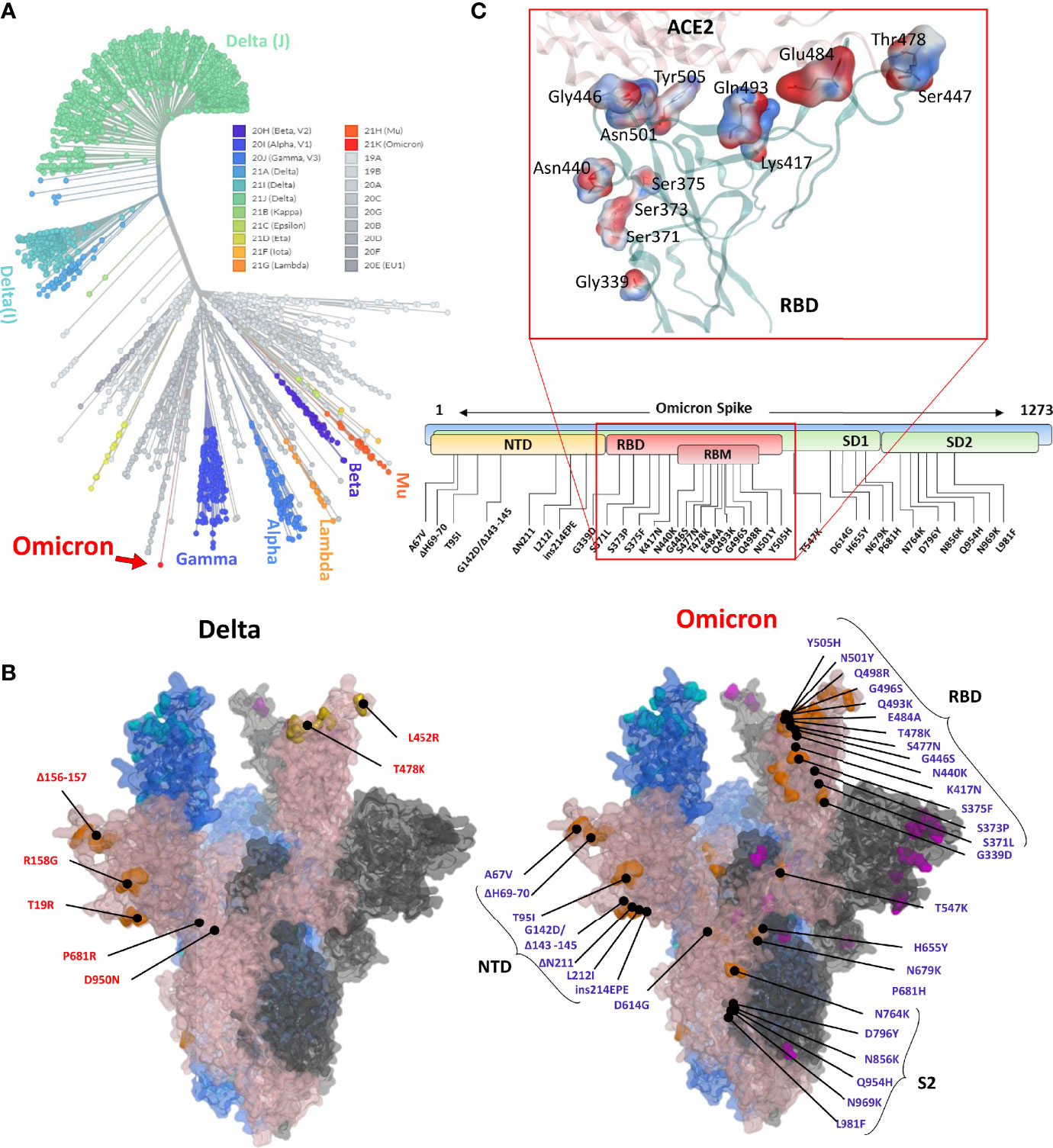
Frontiers | Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies

SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses - ScienceDirect

The displacement of the SARS-CoV-2 variant Delta with Omicron: An investigation of hospital admissions and upper respiratory viral loads - eBioMedicine

Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles | PNAS
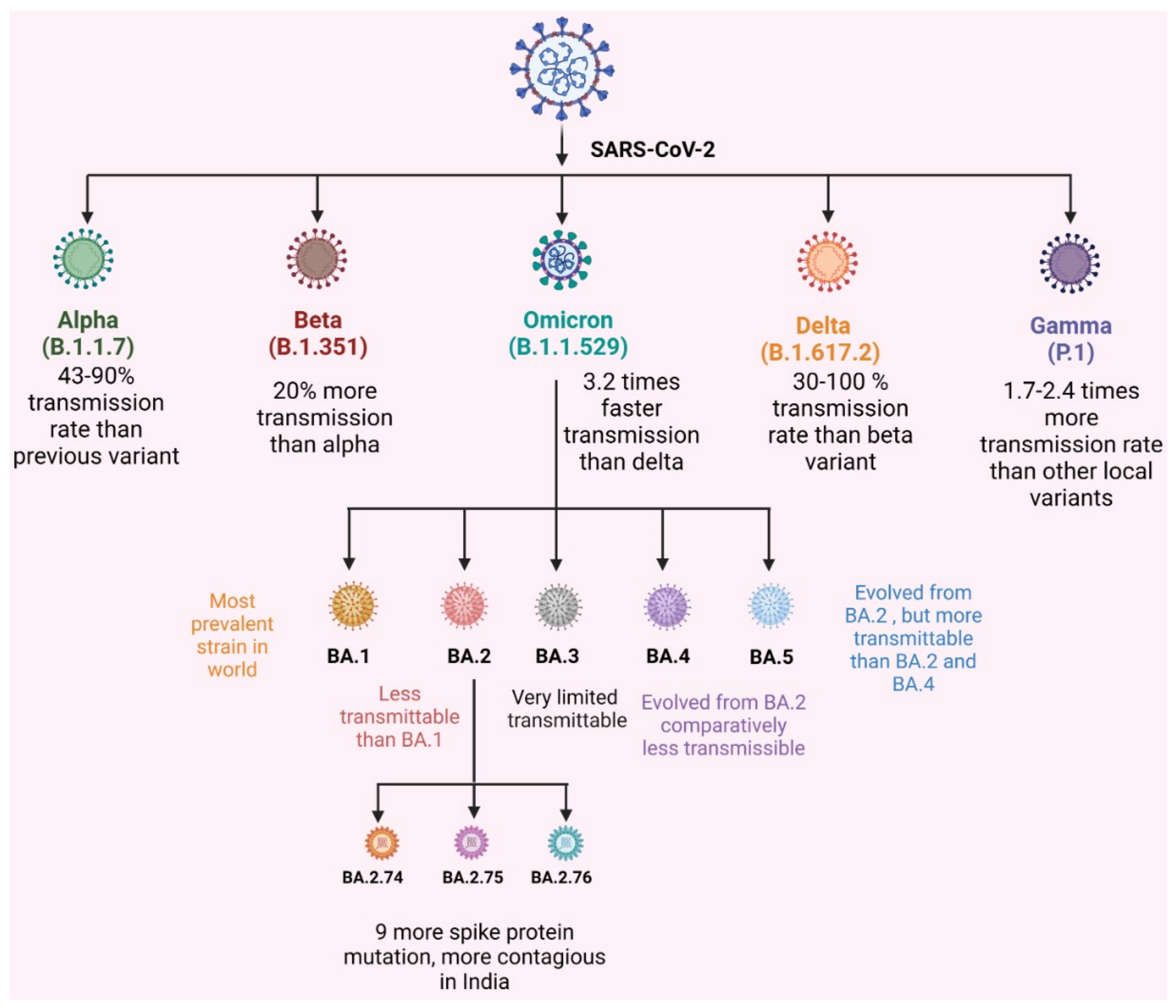
Vaccines | Free Full-Text | Omicron Variant of SARS-CoV-2: An Indian Perspective of Vaccination and Management

Development of a One-Step Qualitative RT-PCR Assay to Detect the SARS-CoV-2 Omicron (B.1.1.529) Variant in Respiratory Specimens | Journal of Clinical Microbiology

Omicron variant of SARS‐CoV‐2: Genomics, transmissibility, and responses to current COVID‐19 vaccines - Araf - 2022 - Journal of Medical Virology - Wiley Online Library
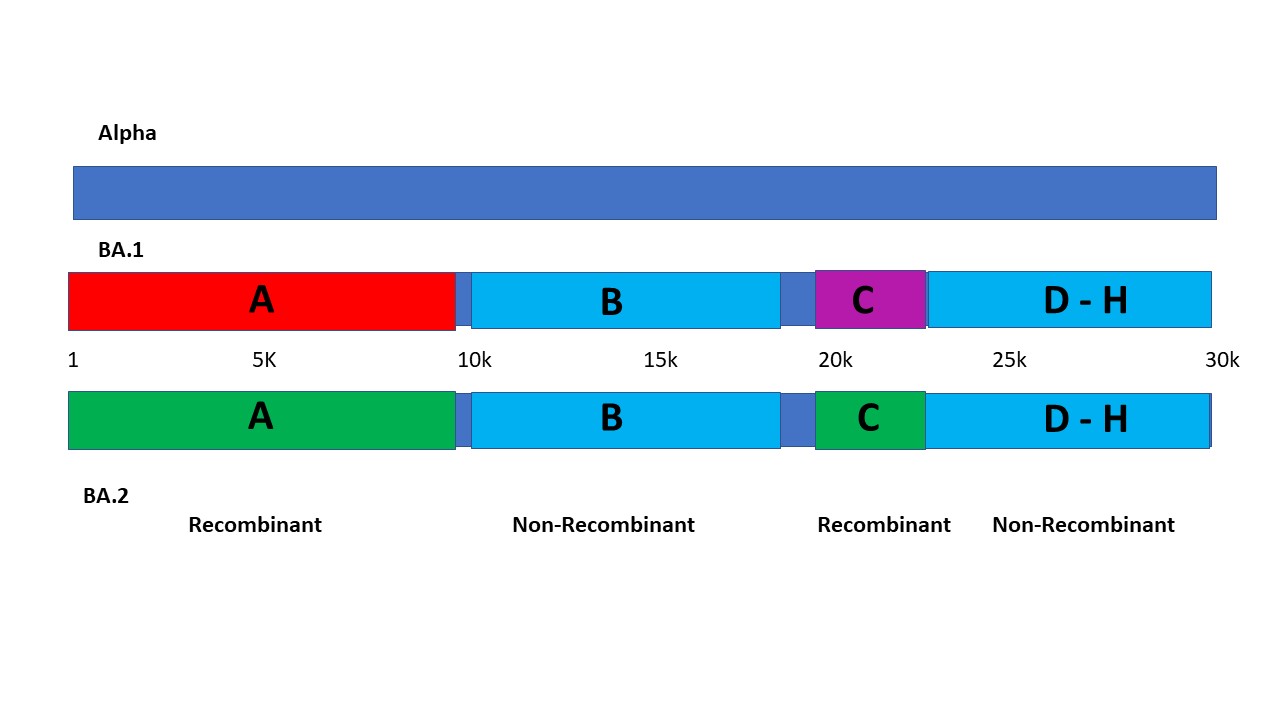
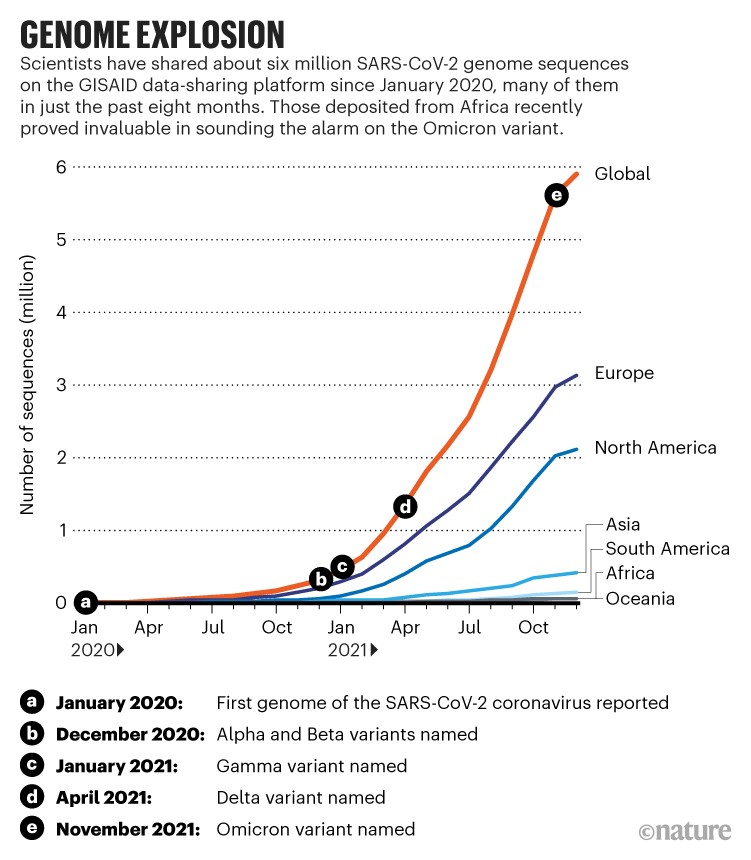
Omicron is a Multiply Recombinant Set of Variants That Have Evolved Over Many Months - SARS-CoV-2 Molecular Evolution - Virological