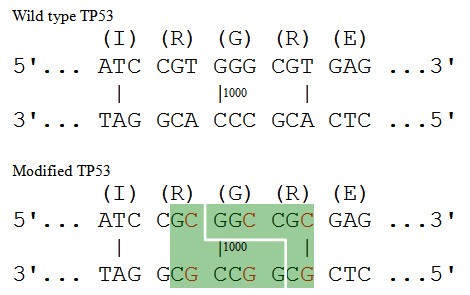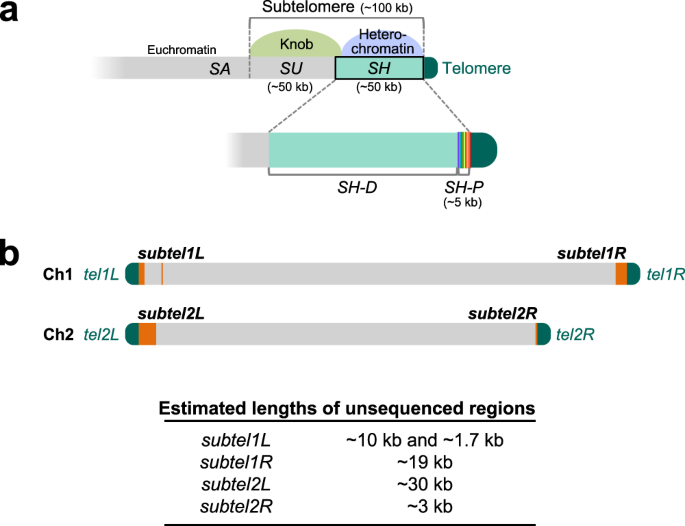
Complete sequences of Schizosaccharomyces pombe subtelomeres reveal multiple patterns of genome variation | Nature Communications

Table 2 from Engineering a rare-cutting restriction enzyme: genetic screening and selection of NotI variants | Semantic Scholar

Sequence characteristics surrounding the NotI site of extra spots and... | Download Scientific Diagram

Molecules | Free Full-Text | Selecting Nanobodies Specific for the Epidermal Growth Factor from a Synthetic Nanobody Library

SOLVED: Based on the DNA sequence given below and Table 2, (the student onlv needs to answer the restriction enzyme (ONE ONLY) CATATGGTAGCGGCCGCATATGTCTAGAAGCGGCCGCTCTAGAG GTATACCATCGCCGGCGTA TACAGATCTTCGCCGGCGAGATCTC Table 2 Restriction Enzyme Noti ...

SOLVED: The restriction enzyme A l u I cleaves at the sequence 5^'- AGCT-3', and NotI cleaves at 5^'- GCGGCCGC-3'. What would be the average distance between cleavage sites for each enzyme
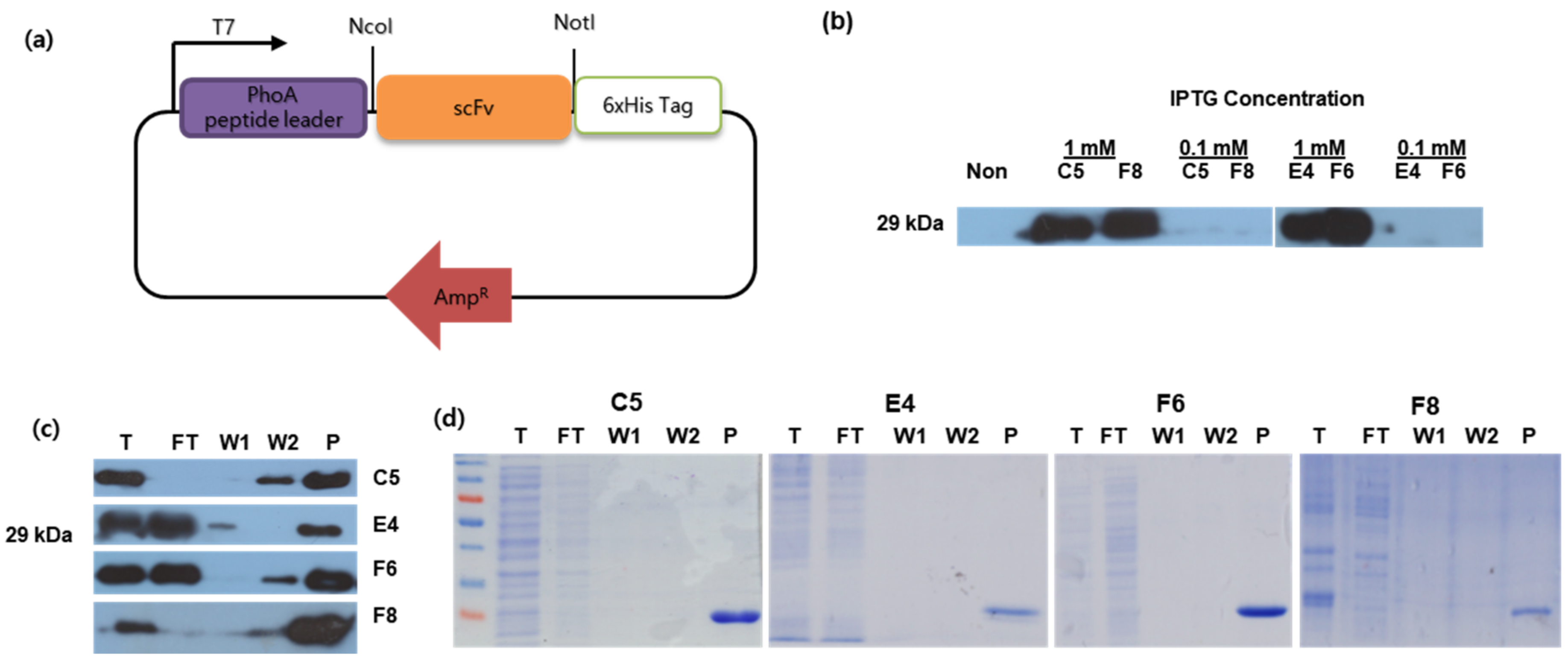
Fishes | Free Full-Text | Development of an Immunoassay Detection System for Koi Herpesvirus Using Recombinant Single-Chain Variable Fragments