NLS vs MMR Dream11 Prediction With Stats, Pitch Report & Player Record of ACPL Kenya T20 Cup, 2022 For Match 3 - ProBatsman

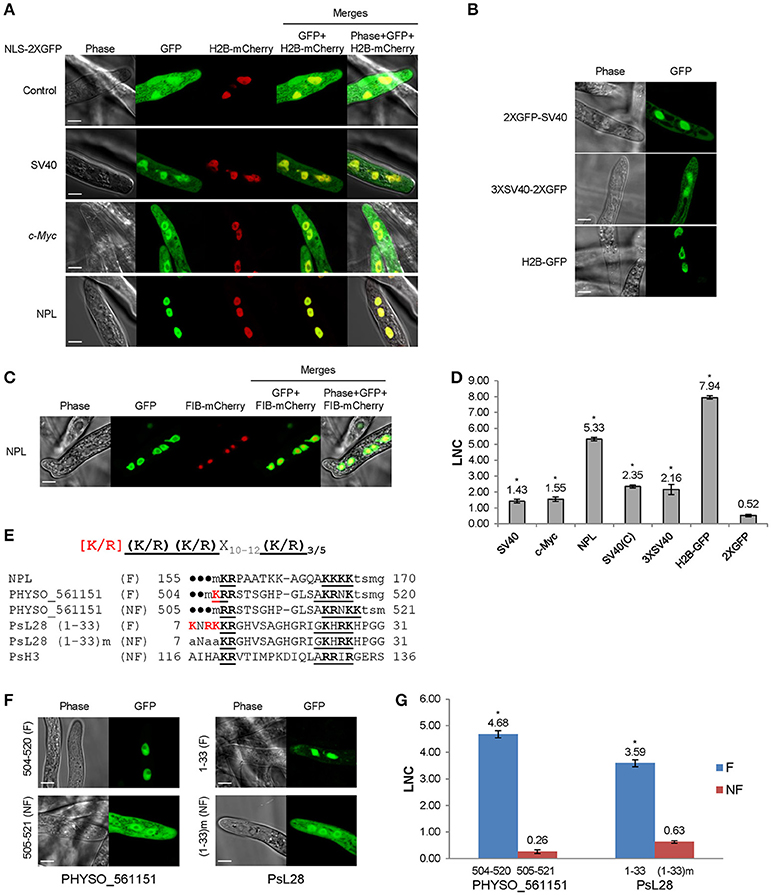
Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells | Nature Communications

Identification of a bipartite nuclear localization signal in the silkworm Masc protein - Sugano - 2016 - FEBS Letters - Wiley Online Library

Prediction results for putative NLS, NES and DNA-binding motifs on CAV... | Download Scientific Diagram

Cells | Free Full-Text | Bioinformatics and Functional Analysis of a New Nuclear Localization Sequence of the Influenza A Virus Nucleoprotein
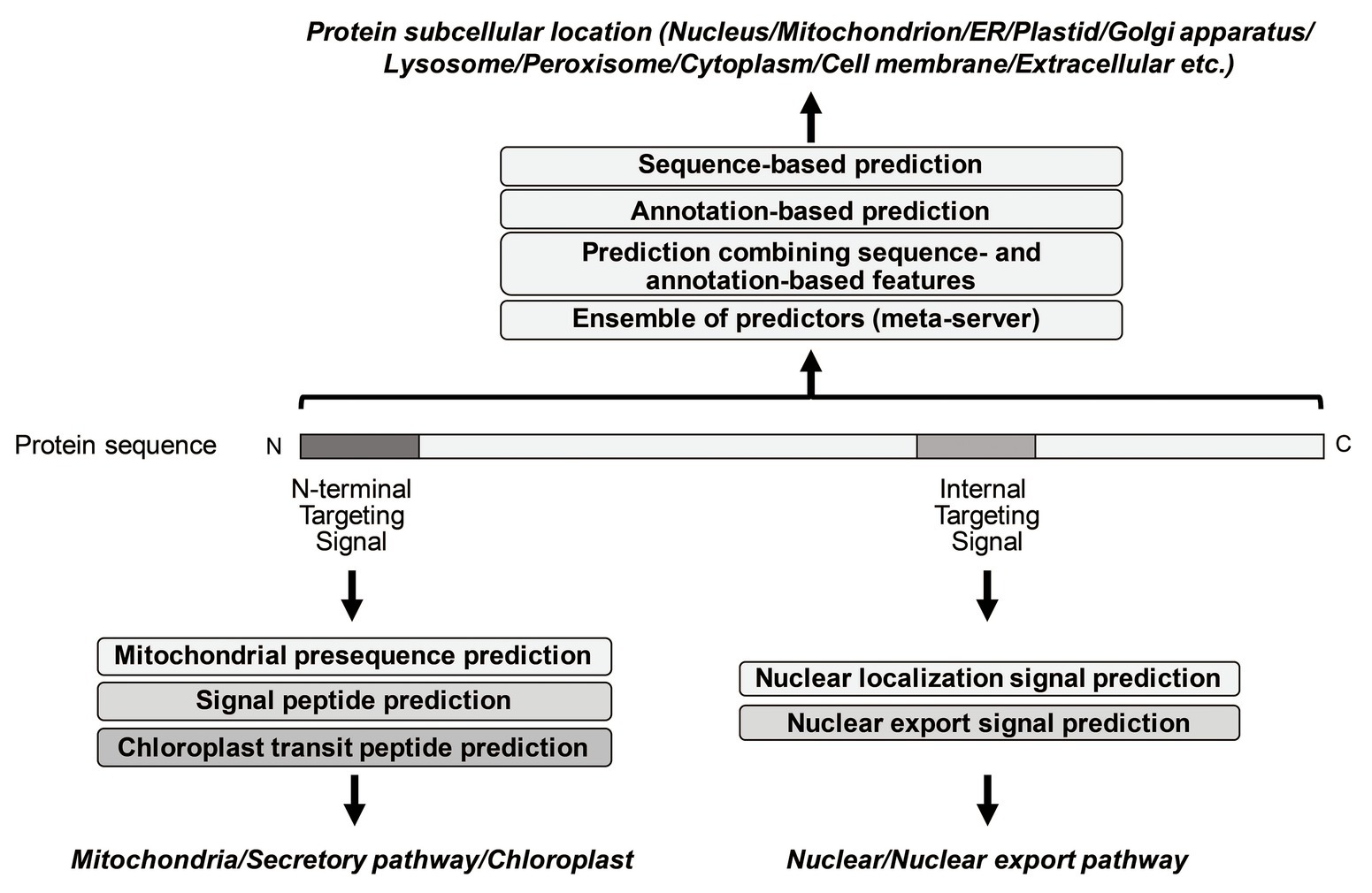
Frontiers | Tools for the Recognition of Sorting Signals and the Prediction of Subcellular Localization of Proteins From Their Amino Acid Sequences
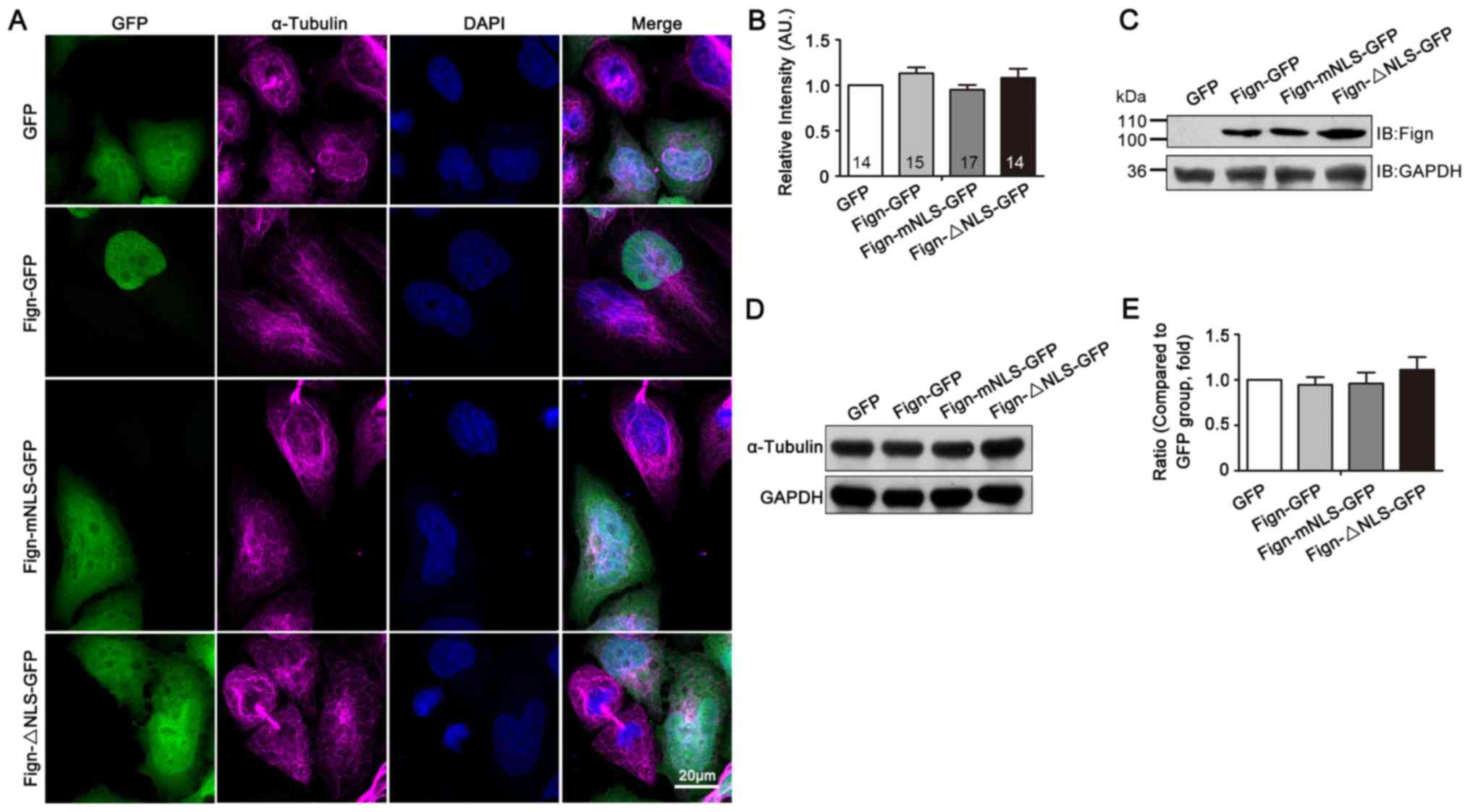
A nuclear localization signal is required for the nuclear translocation of Fign and its microtubule‑severing function
SeqNLS: Nuclear Localization Signal Prediction Based on Frequent Pattern Mining and Linear Motif Scoring | PLOS ONE

Discovering nuclear targeting signal sequence through protein language learning and multivariate analysis - ScienceDirect

Identification of nuclear localization signal and nuclear export signal of VP1 from the chicken anemia virus and effects on VP2 shuttling in cells | Virology Journal | Full Text

Variations in nuclear localization strategies among pol X family enzymes - Kirby - 2018 - Traffic - Wiley Online Library

Potential NLS sequences in TPs of Streptomyces. (A) The archetypal Tpg... | Download Scientific Diagram

Crystallographic Analysis of the Recognition of a Nuclear Localization Signal by the Nuclear Import Factor Karyopherin α: Cell
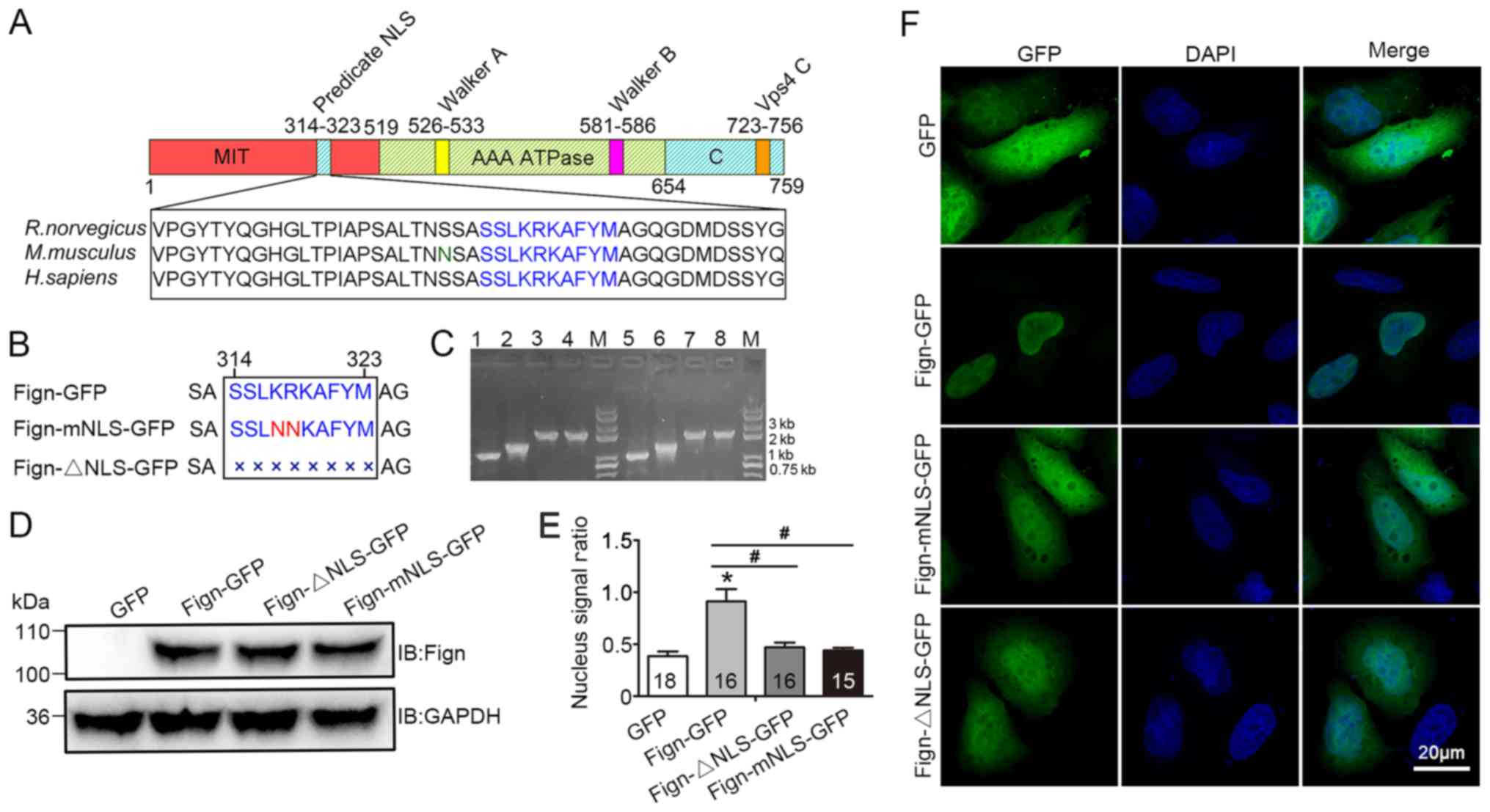
A nuclear localization signal is required for the nuclear translocation of Fign and its microtubule‑severing function
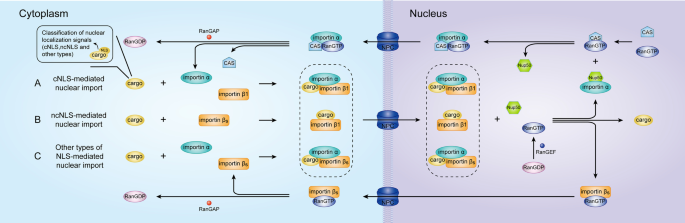
Types of nuclear localization signals and mechanisms of protein import into the nucleus | Cell Communication and Signaling | Full Text