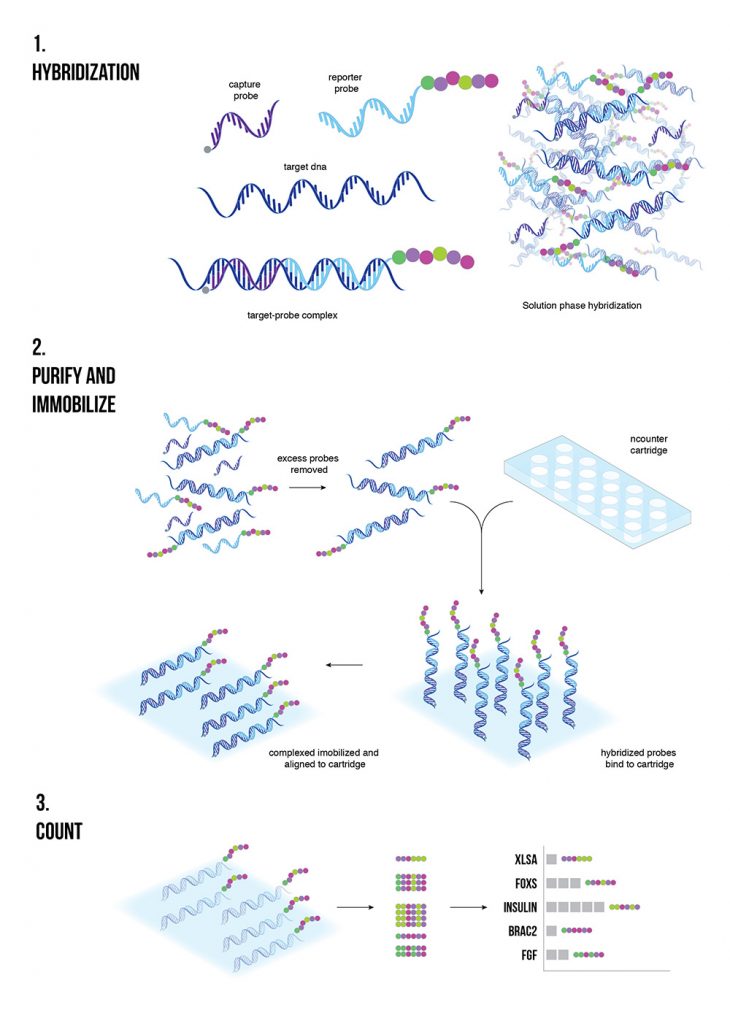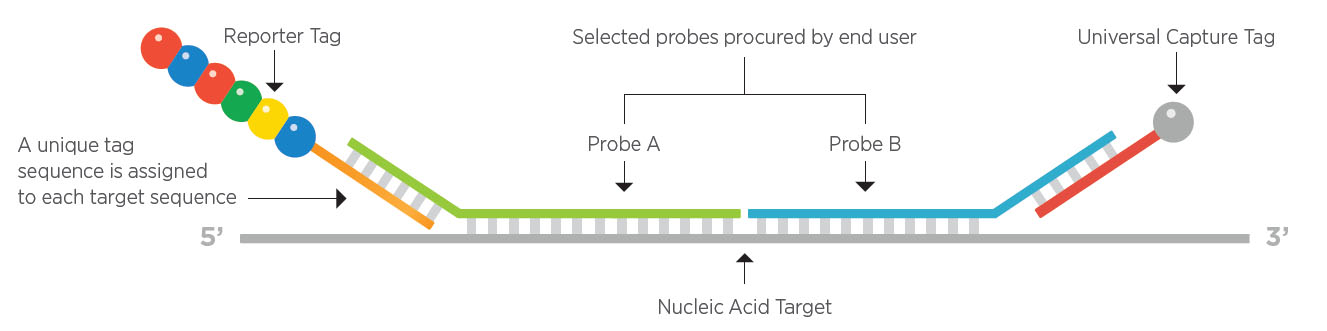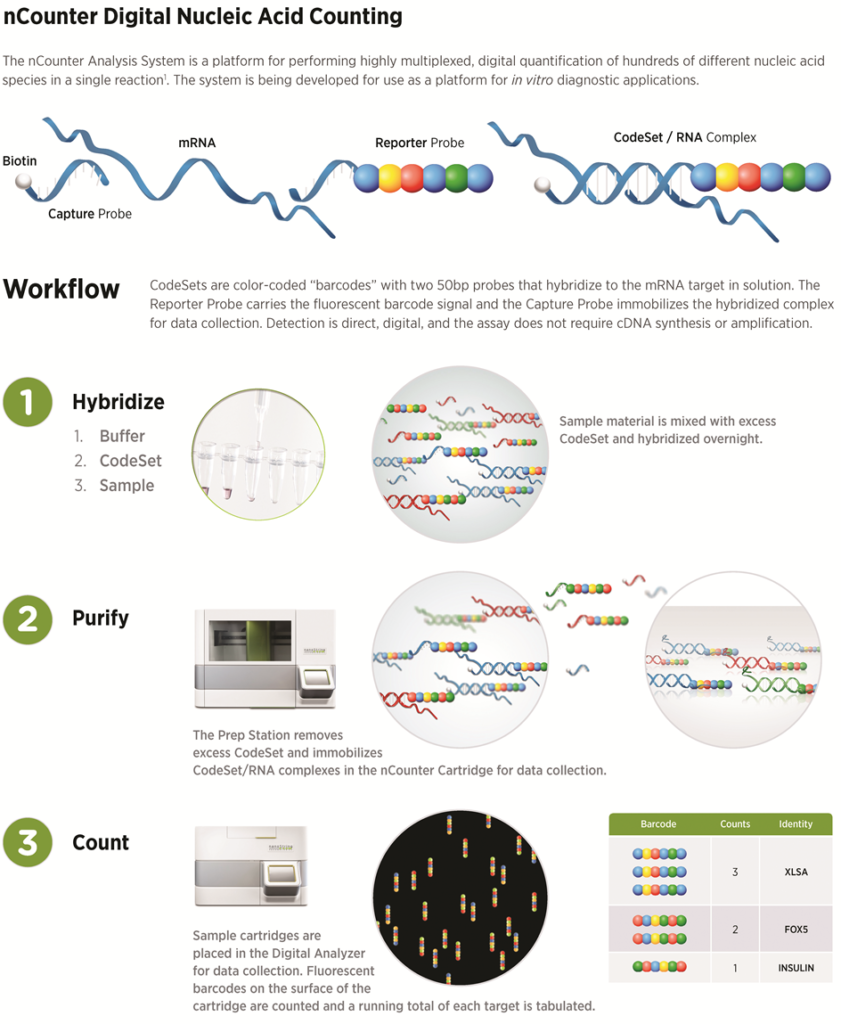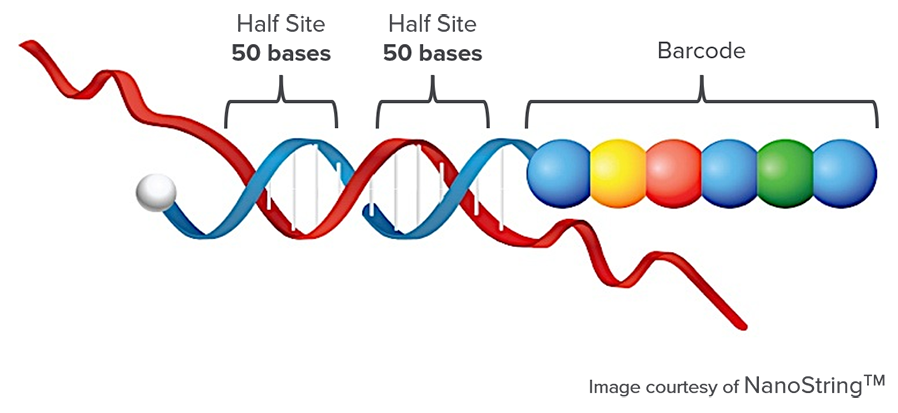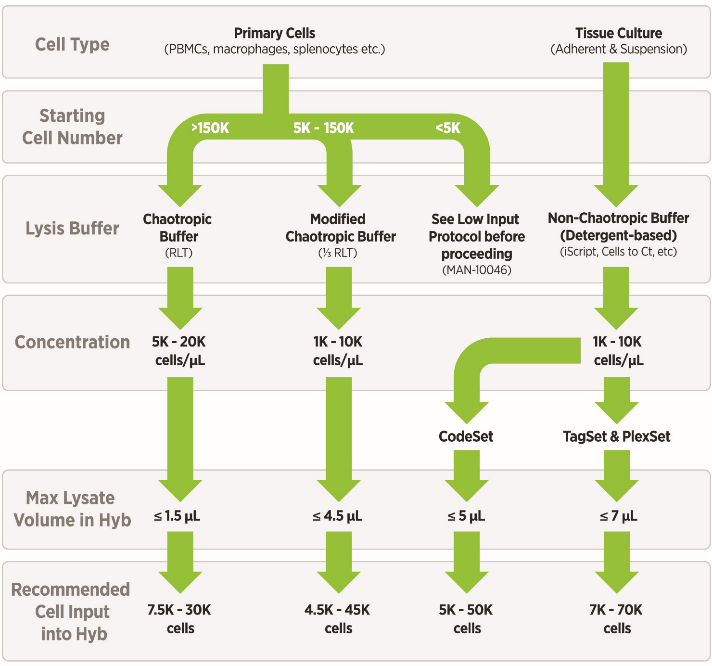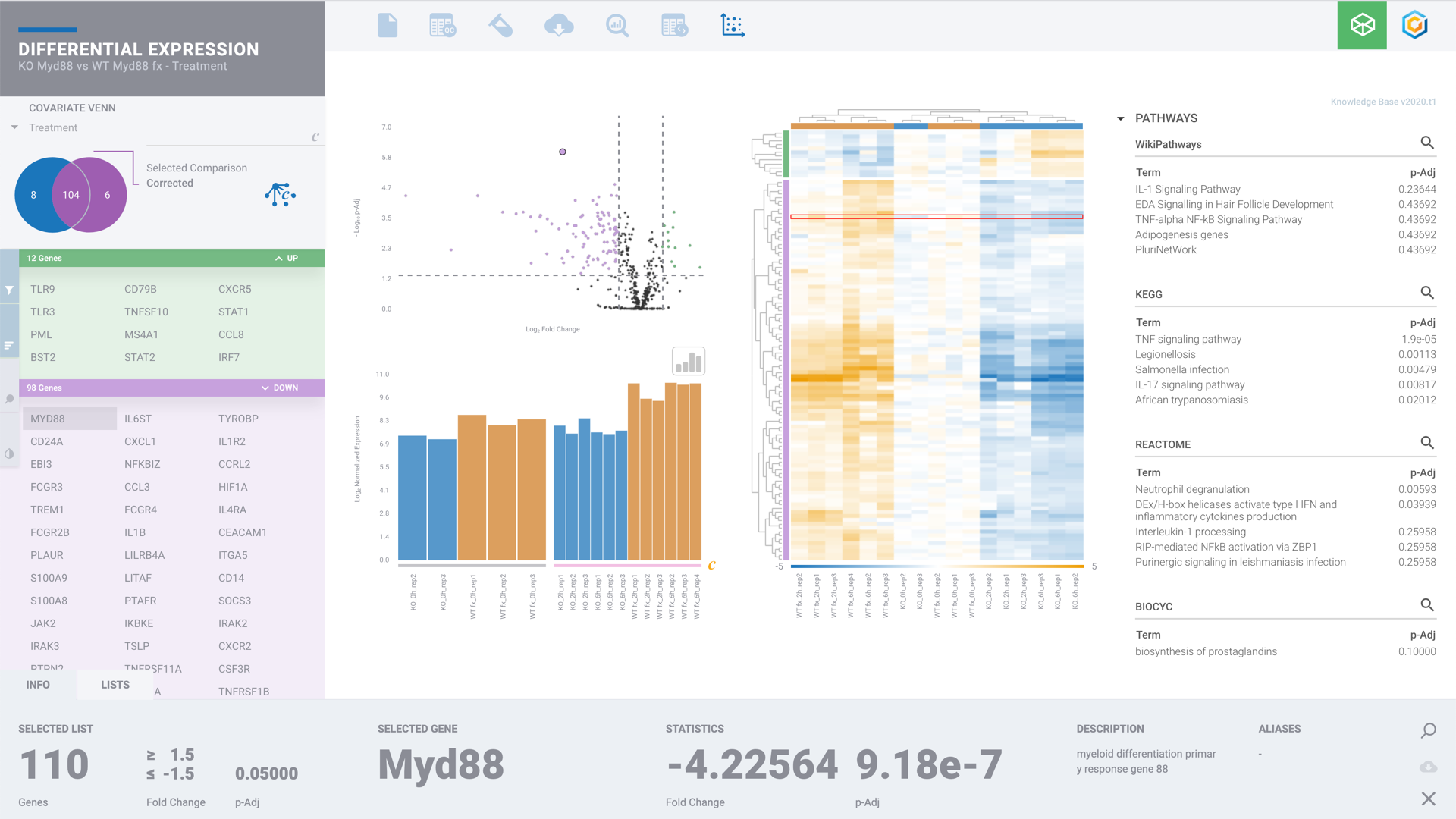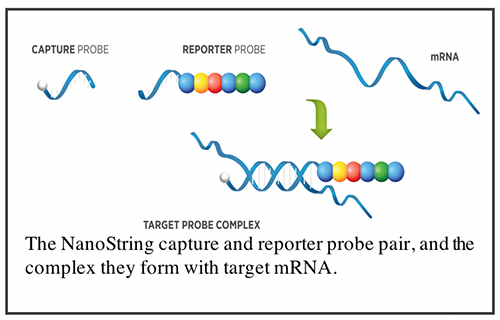
NanoString for Highly Multiplexed Target Gene Analysis: Center for Genetic Medicine: Feinberg School of Medicine: Northwestern University
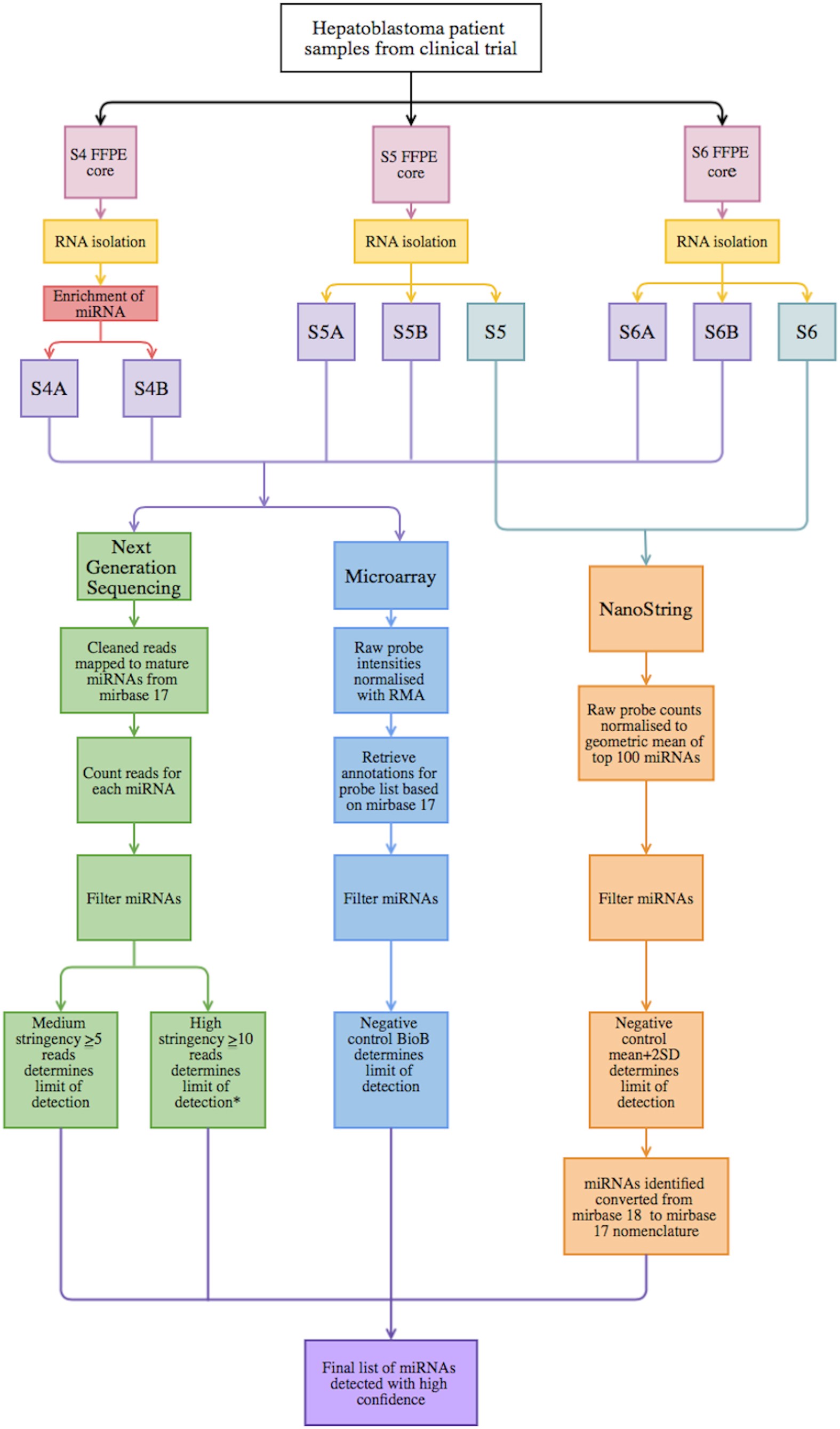
A cross comparison of technologies for the detection of microRNAs in clinical FFPE samples of hepatoblastoma patients | Scientific Reports
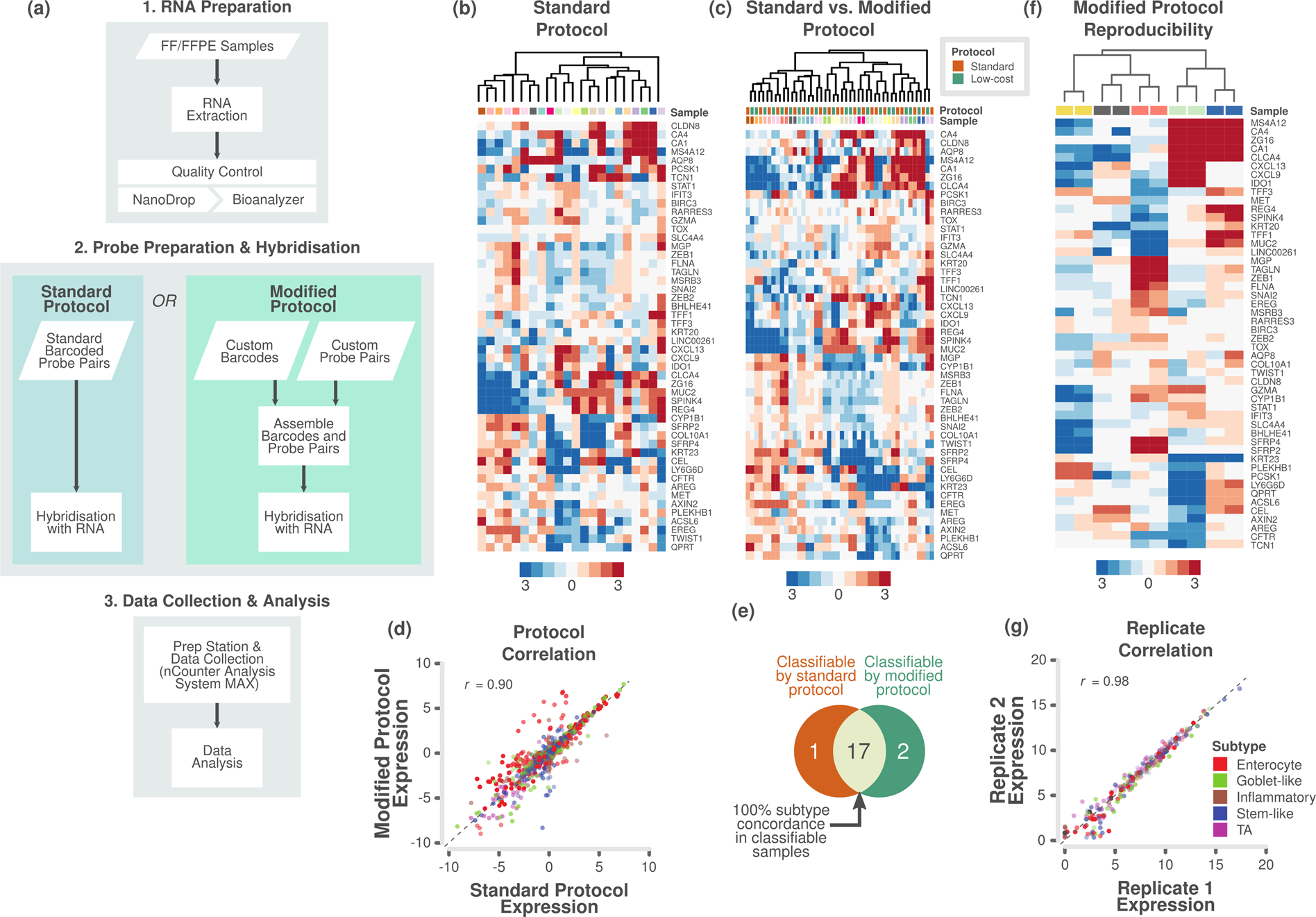
Analytical Validation of Multiplex Biomarker Assay to Stratify Colorectal Cancer into Molecular Subtypes | Scientific Reports
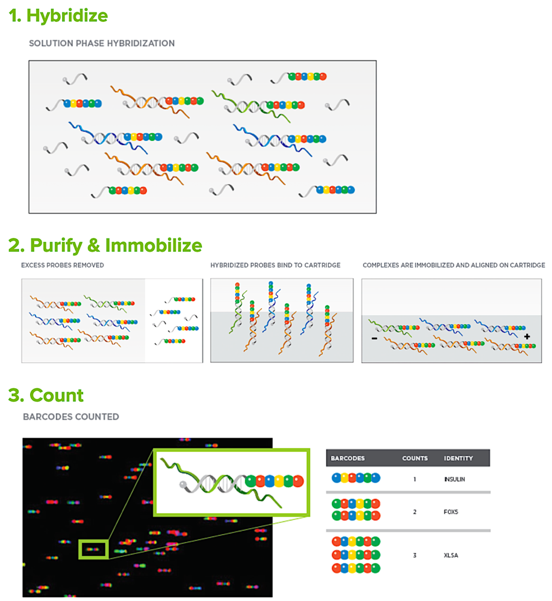
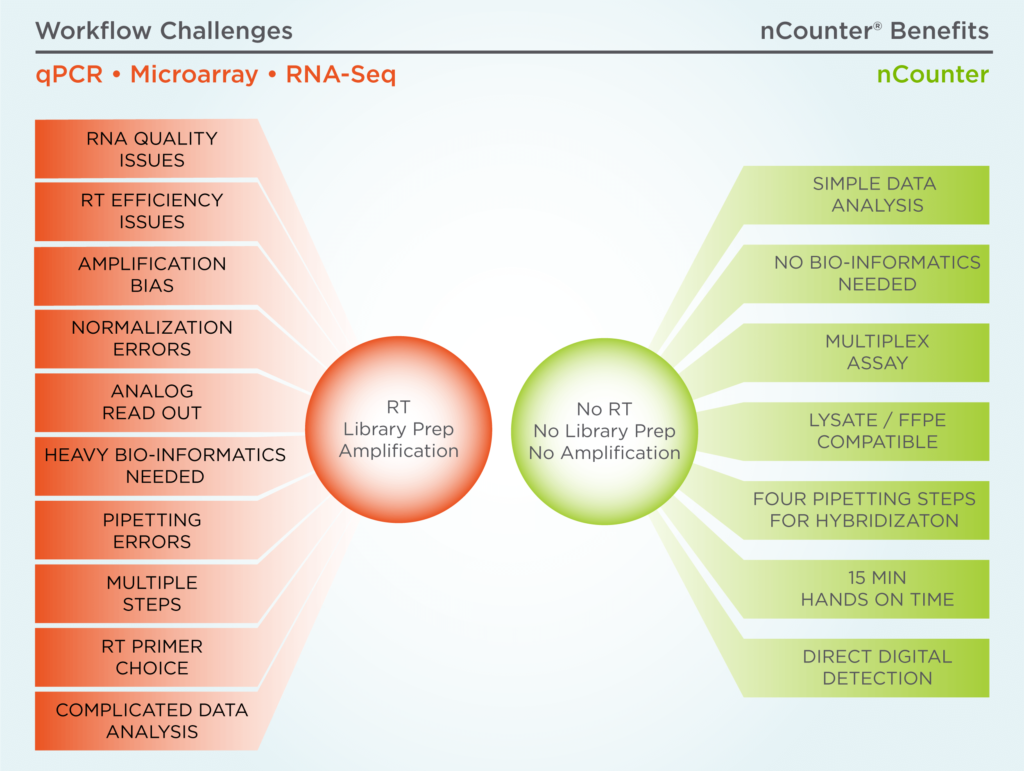
NanoString Resources for Quantitative Analysis of Gene Expression – Resource for Developmental Regulatory Genomics

Reference-free transcriptome exploration reveals novel RNAs for prostate cancer diagnosis | Life Science Alliance

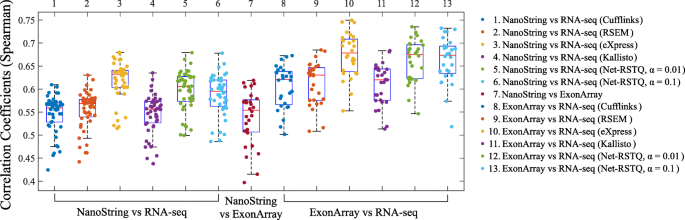
Comparison of RNA-Seq and NanoString data sets. For each gene and time... | Download Scientific Diagram

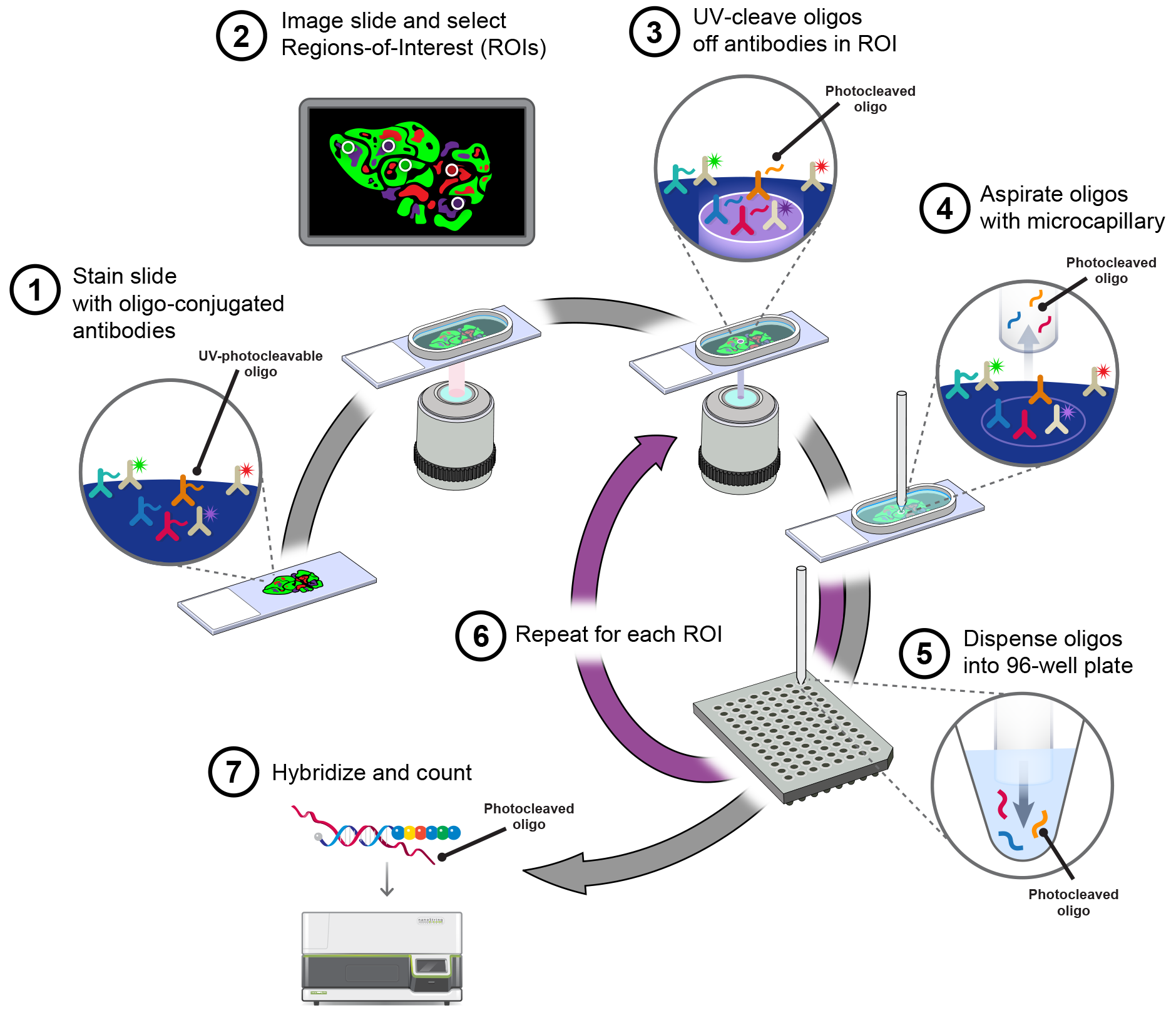
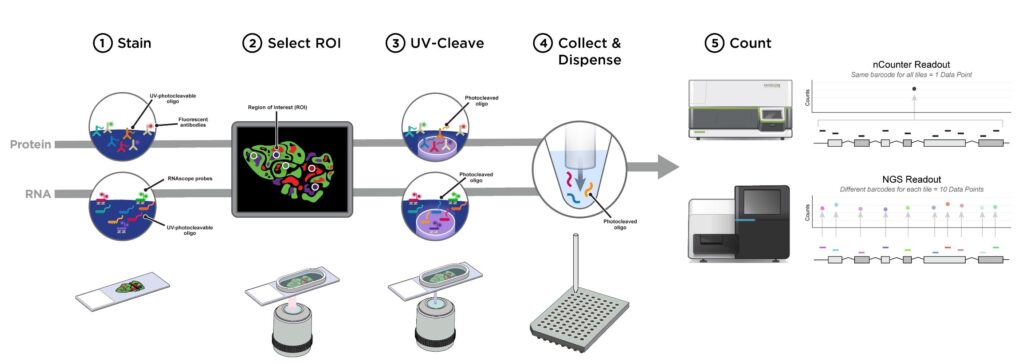
NanoString detection method schematic. Frozen samples are lyophilized,... | Download Scientific Diagram