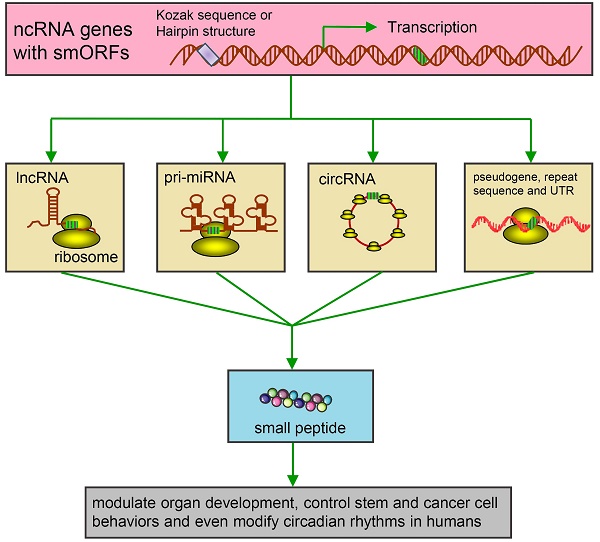
IJMS | Free Full-Text | Kozak Similarity Score Algorithm Identifies Alternative Translation Initiation Codons Implicated in Cancers
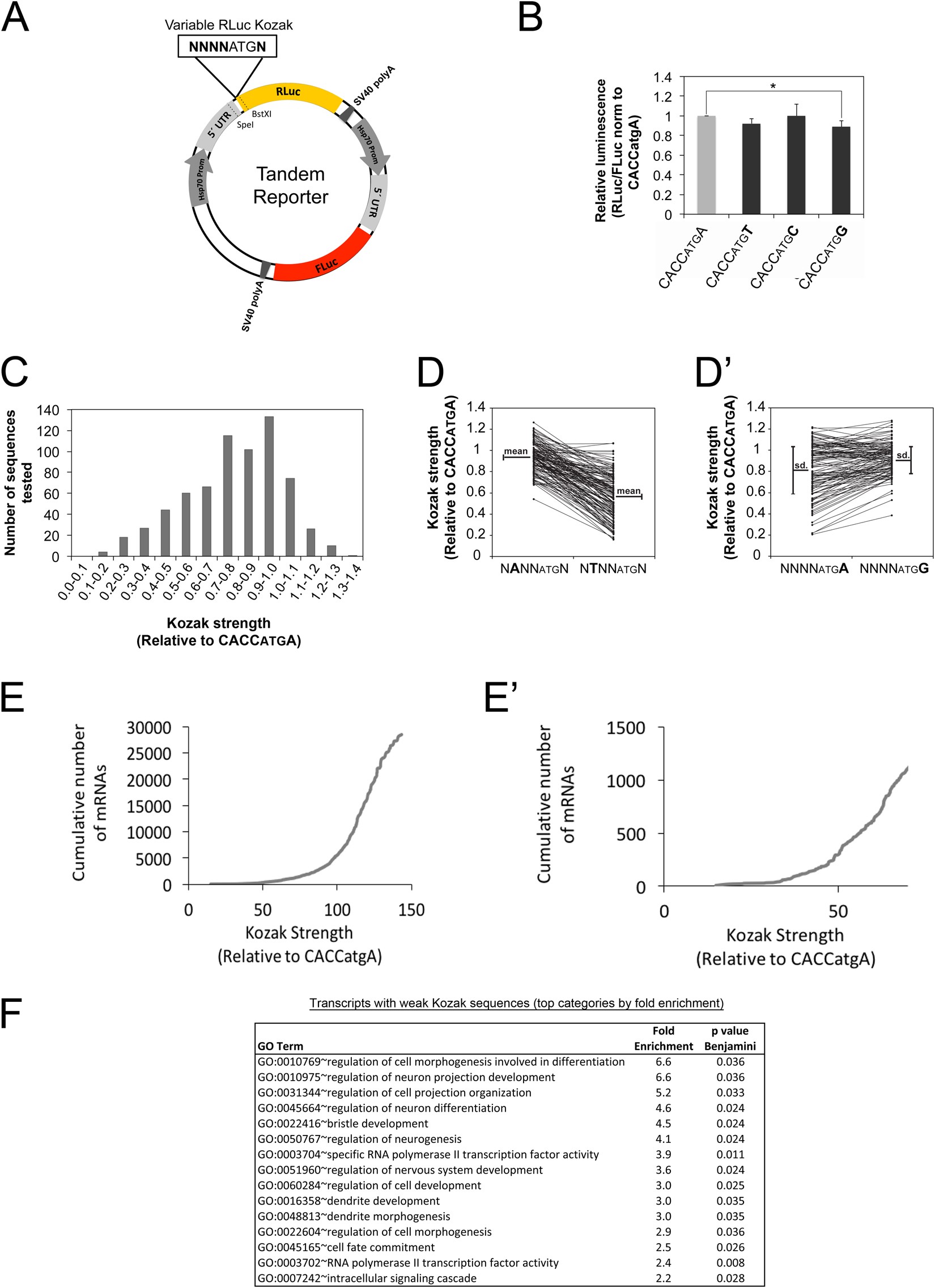
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports
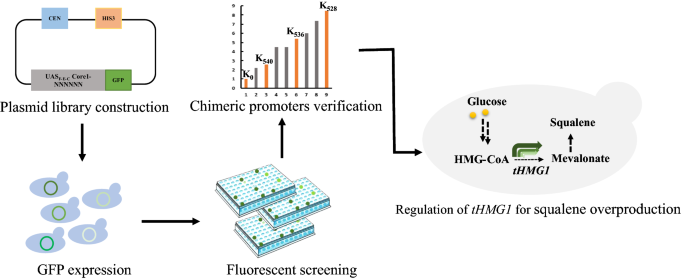
Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants | Microbial Cell Factories | Full Text
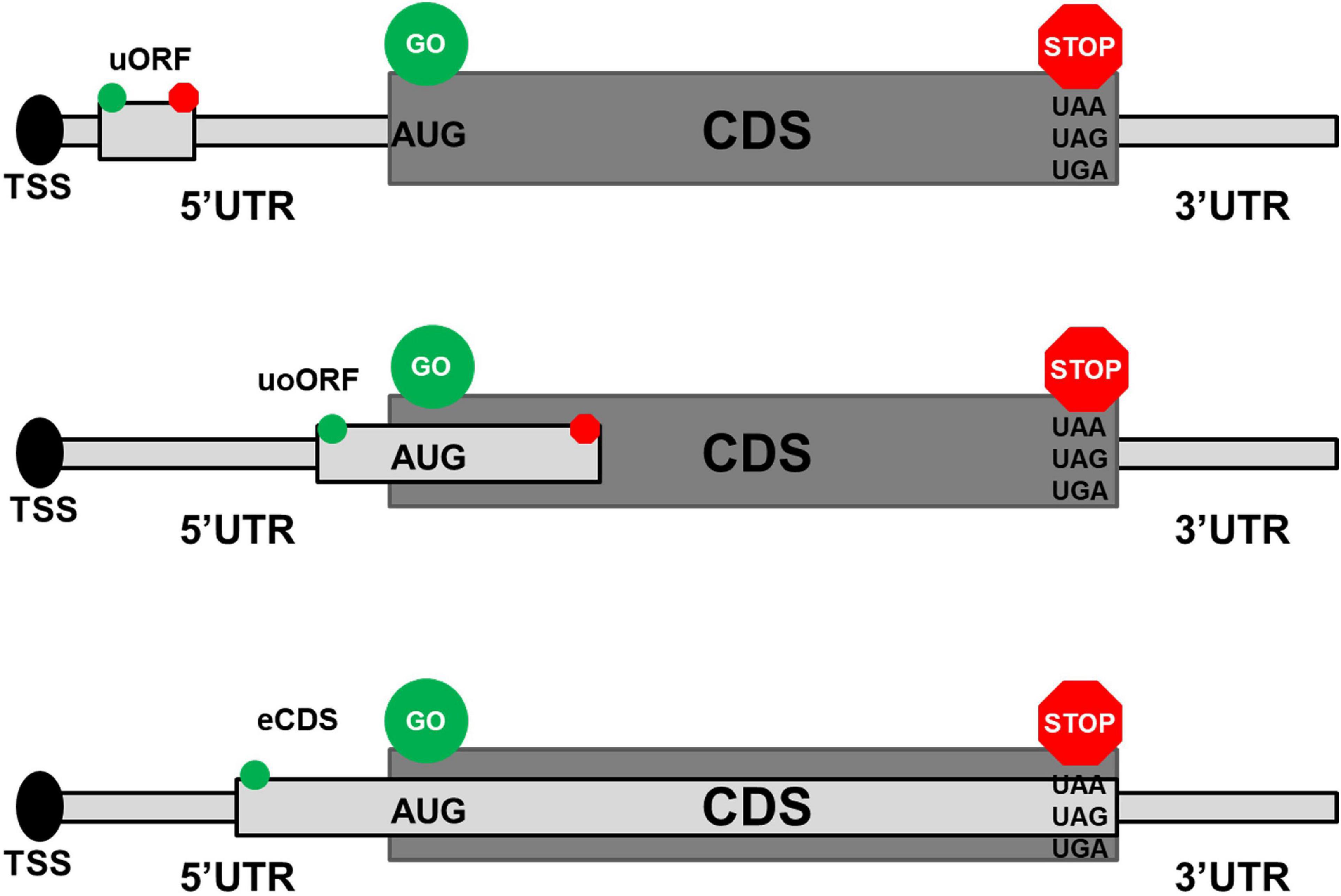
Frontiers | Common and Rare 5′UTR Variants Altering Upstream Open Reading Frames in Cardiovascular Genomics

Tailoring translational strength using Kozak sequence variants improves bispecific antibody assembly and reduces product‐related impurities in CHO cells - Blanco - 2020 - Biotechnology and Bioengineering - Wiley Online Library

Translation of small downstream ORFs enhances translation of canonical main open reading frames | The EMBO Journal
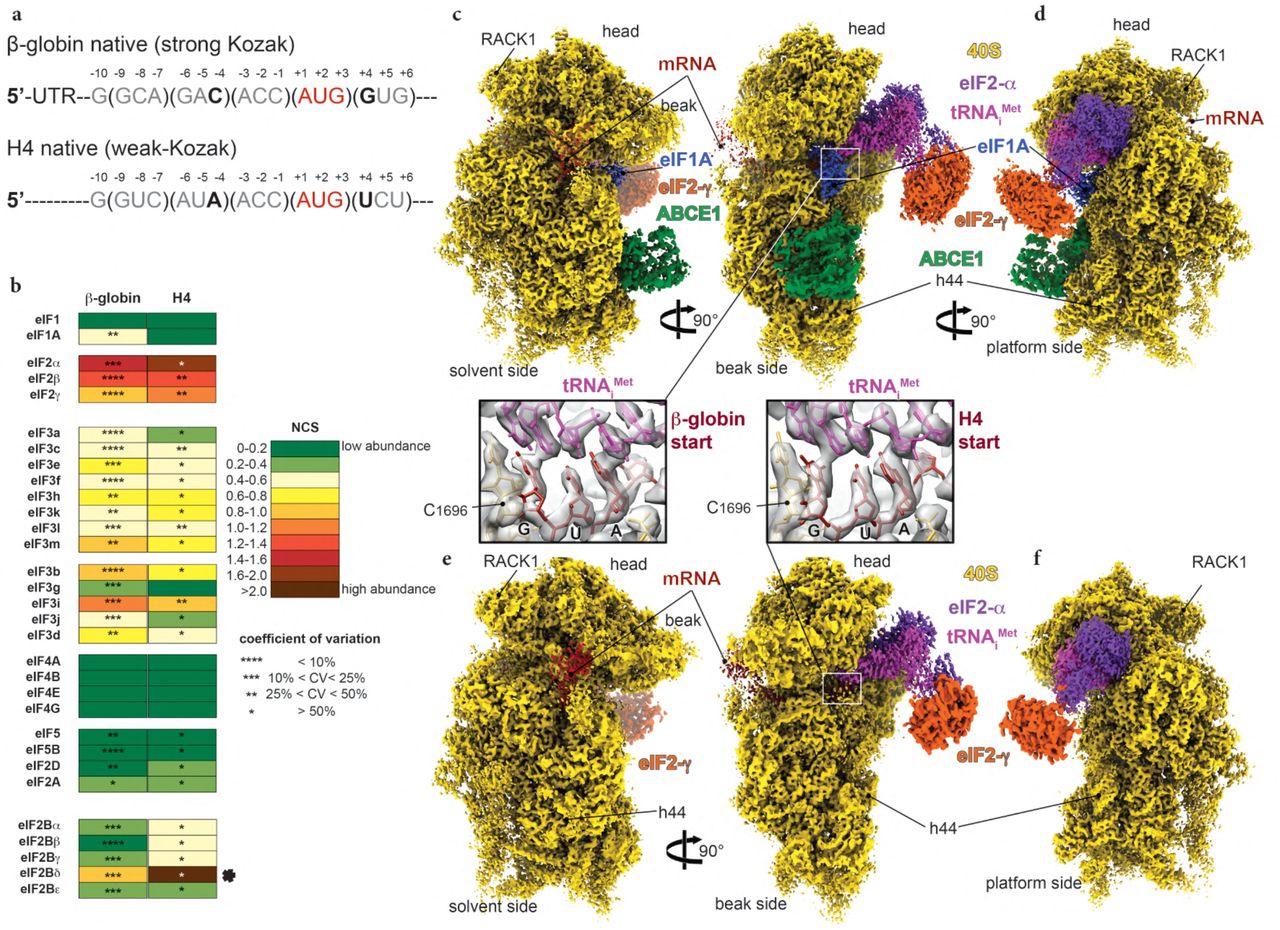
Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv

Analysis of Kozak consensus sequences surrounding the start codon AUG... | Download Scientific Diagram

Rps26 directs mRNA-specific translation by recognition of Kozak sequence elements | Nature Structural & Molecular Biology

Human 5' UTR design and variant effect prediction from a massively parallel translation assay. - Abstract - Europe PMC
Deep learning of the regulatory grammar of yeast 5′ untranslated regions from 500,000 random sequences

Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants | Microbial Cell Factories | Full Text

An expanded toolkit for Drosophila gene tagging using synthesized homology donor constructs for CRISPR mediated homologous recombination | bioRxiv

A non-coding A-to-T Kozak site change related to the transmissibility of Alpha, Delta and Omicron VOCs - Abstract - Europe PMC

Relationship between consensus Kozak sequence at the true start ATG for... | Download Scientific Diagram
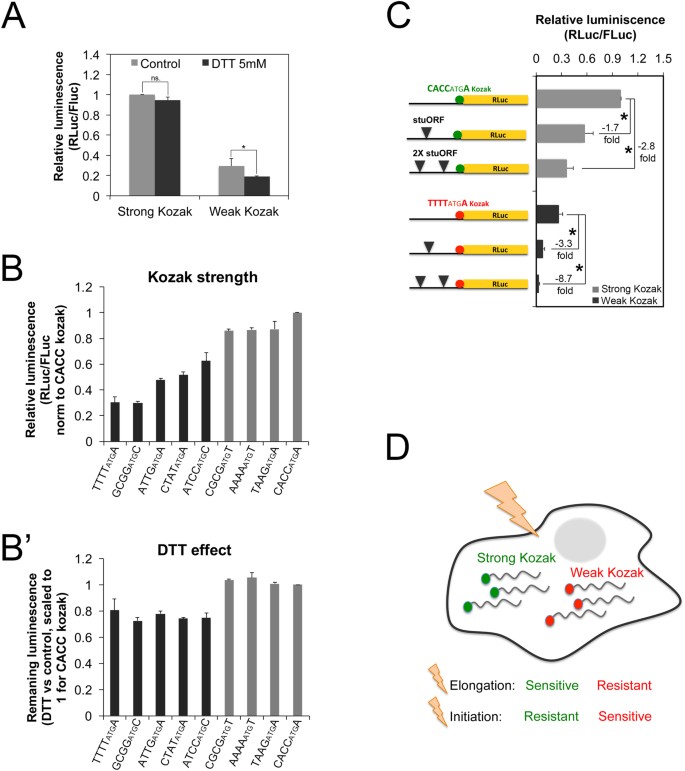
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports