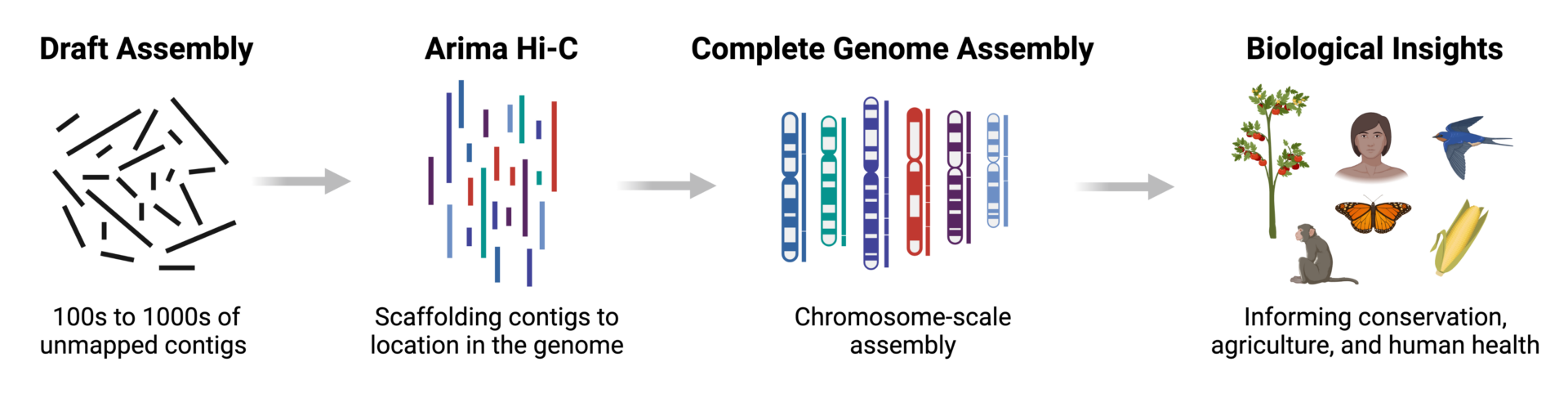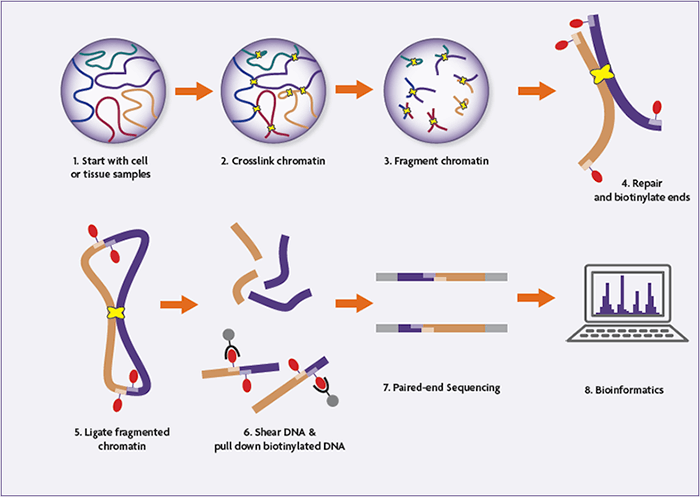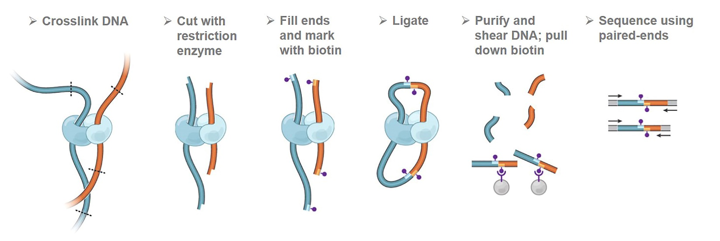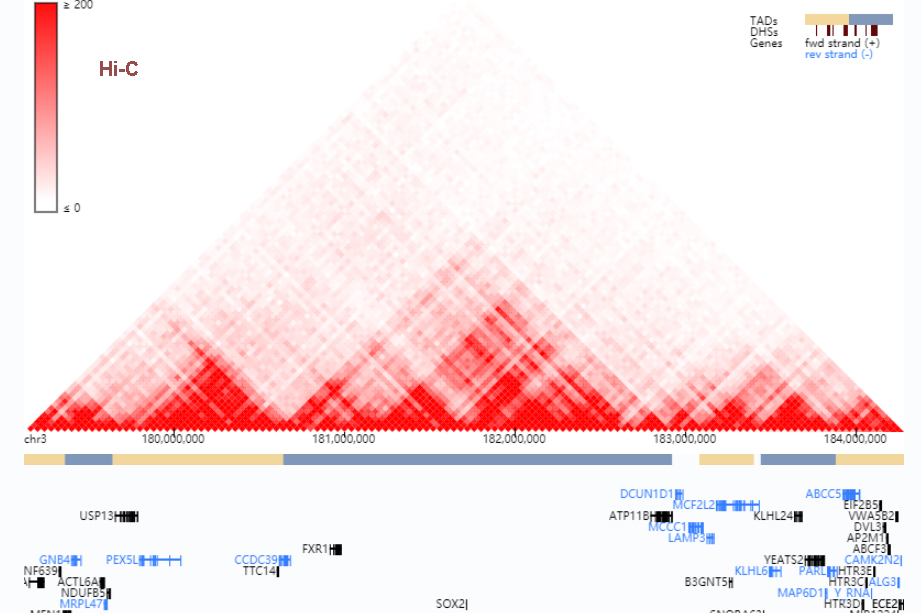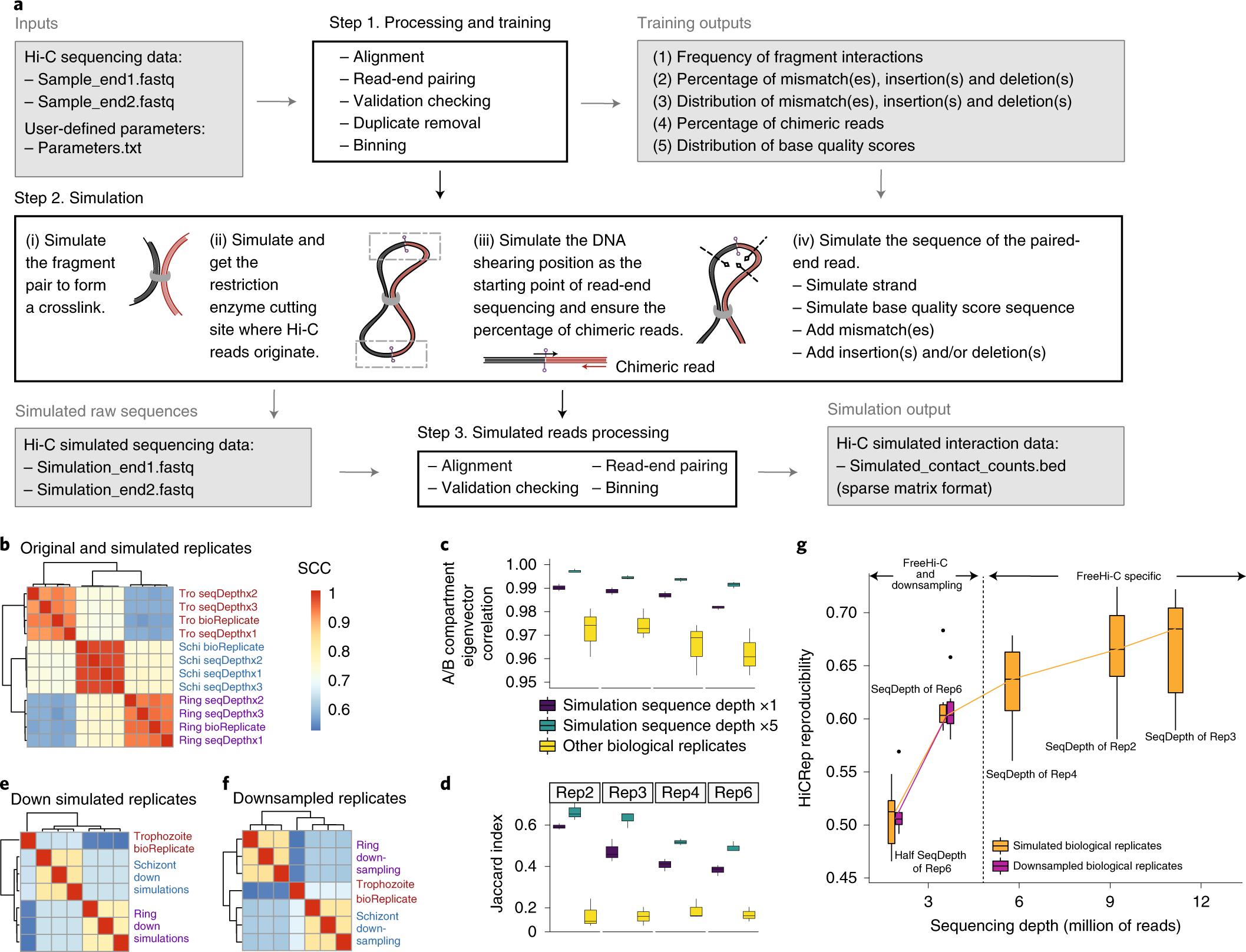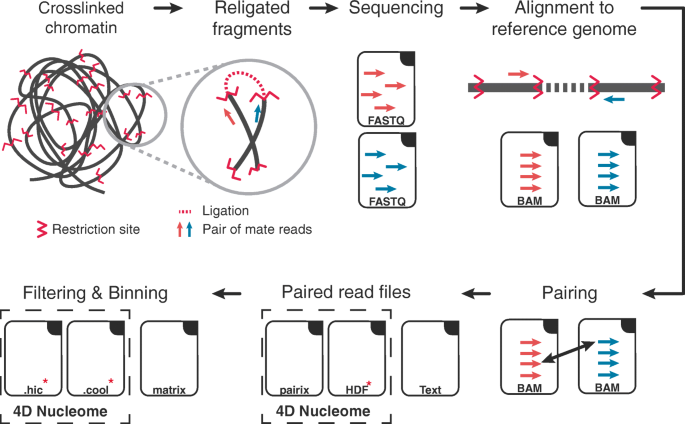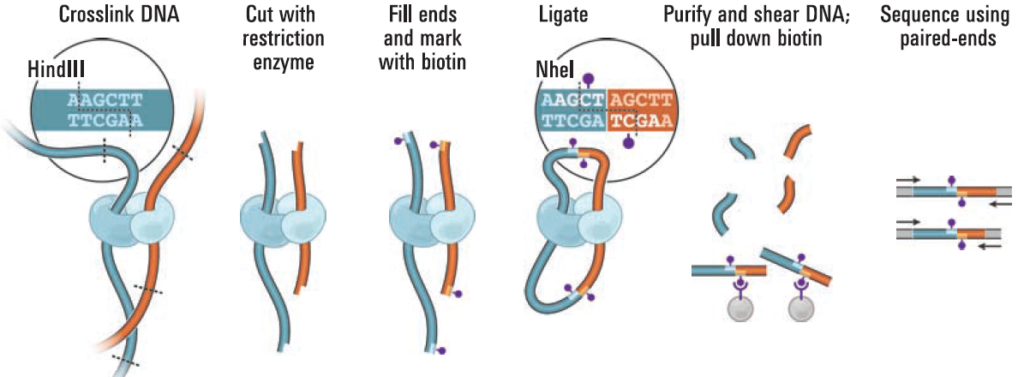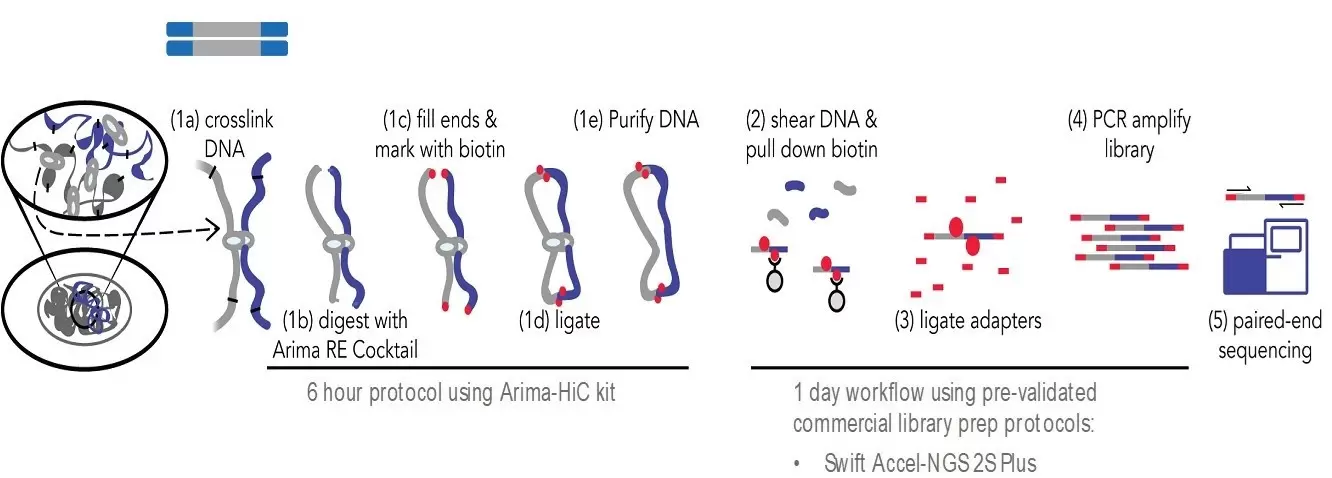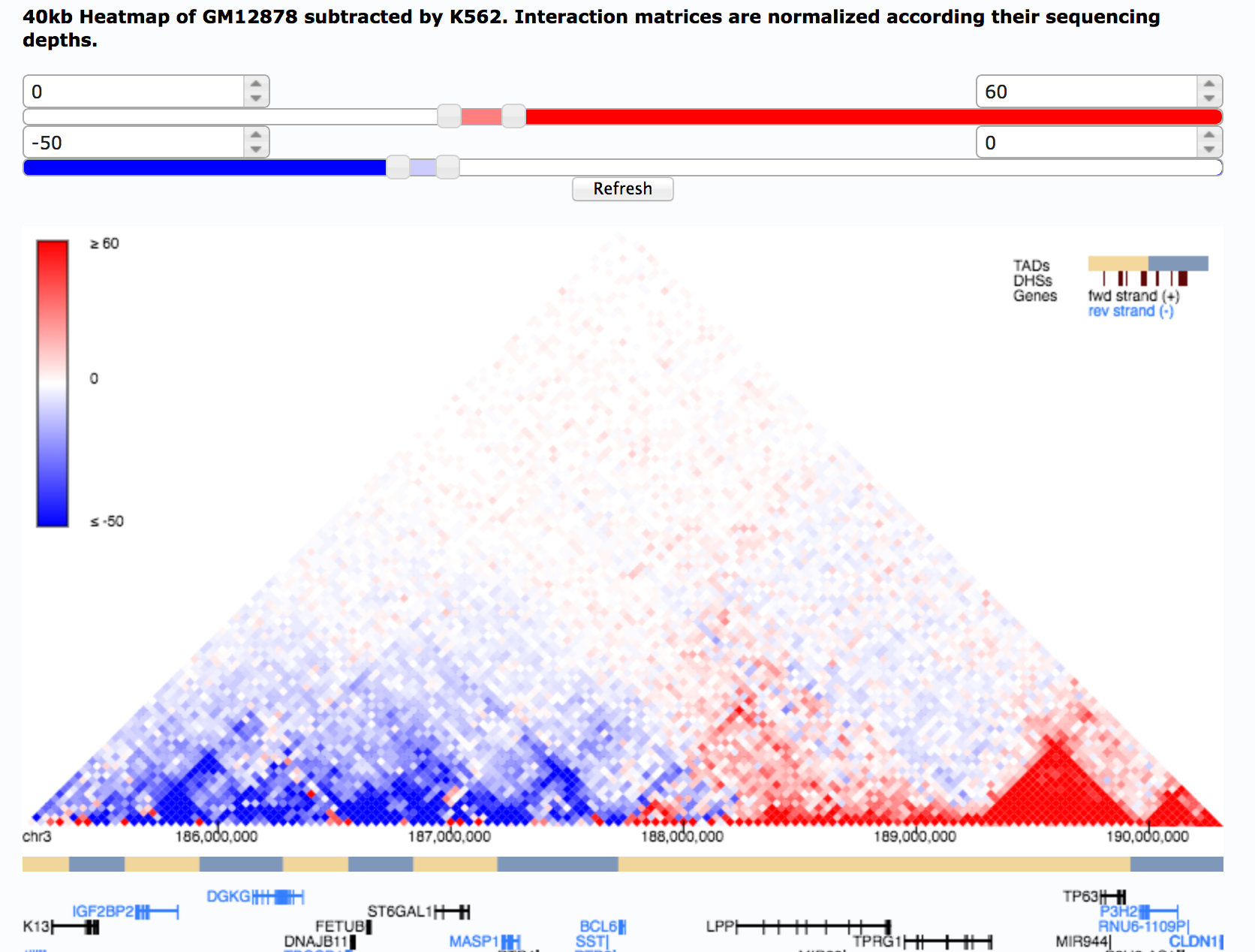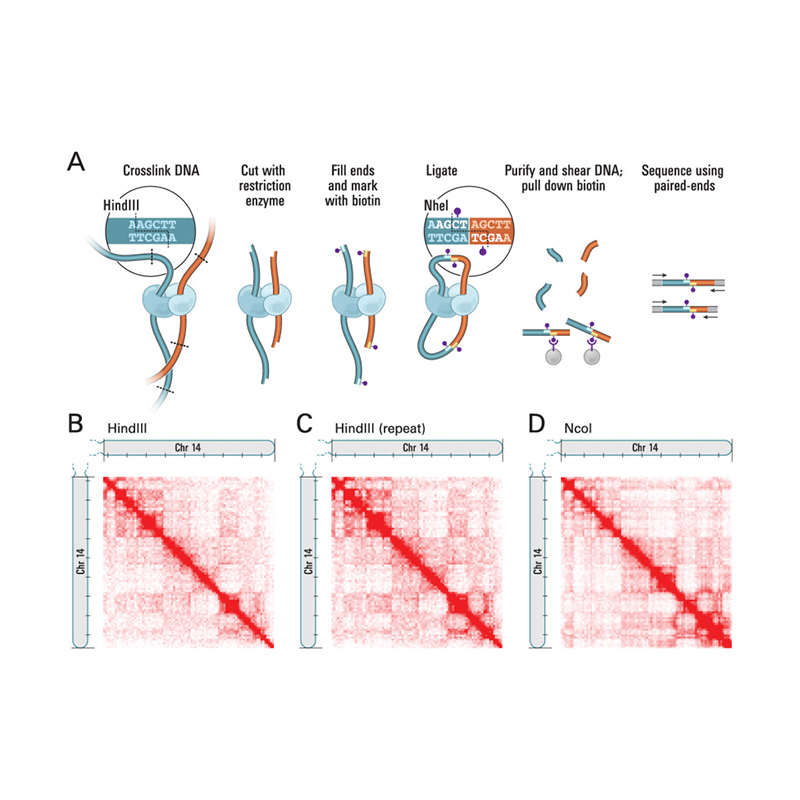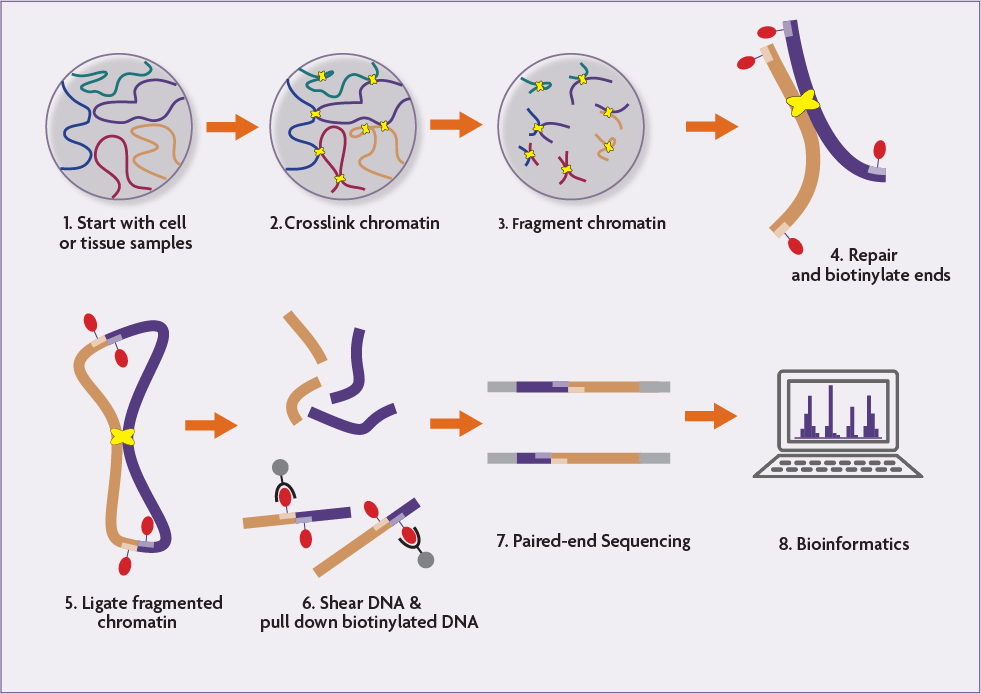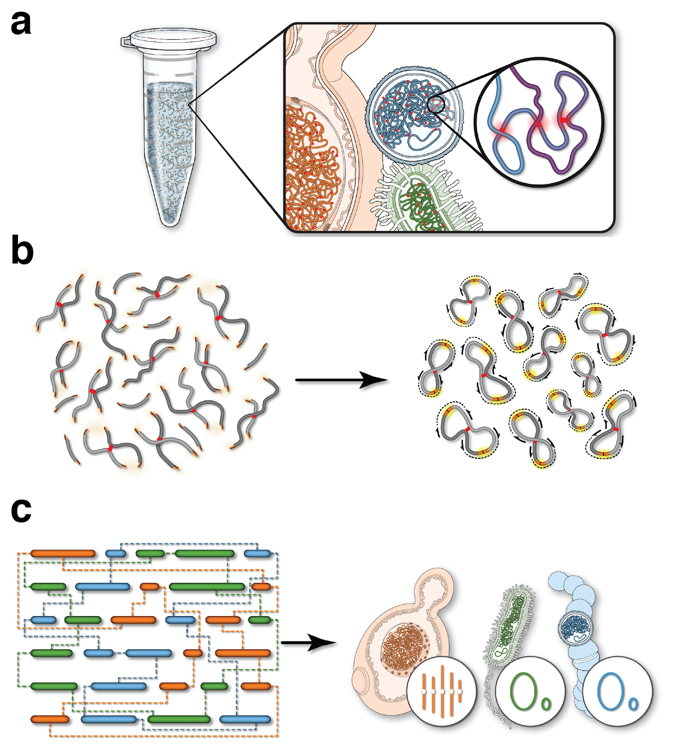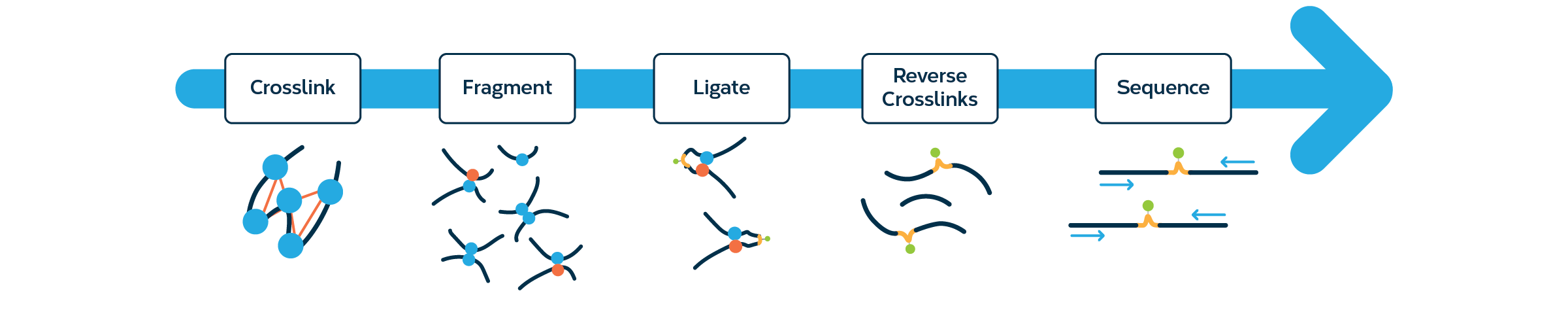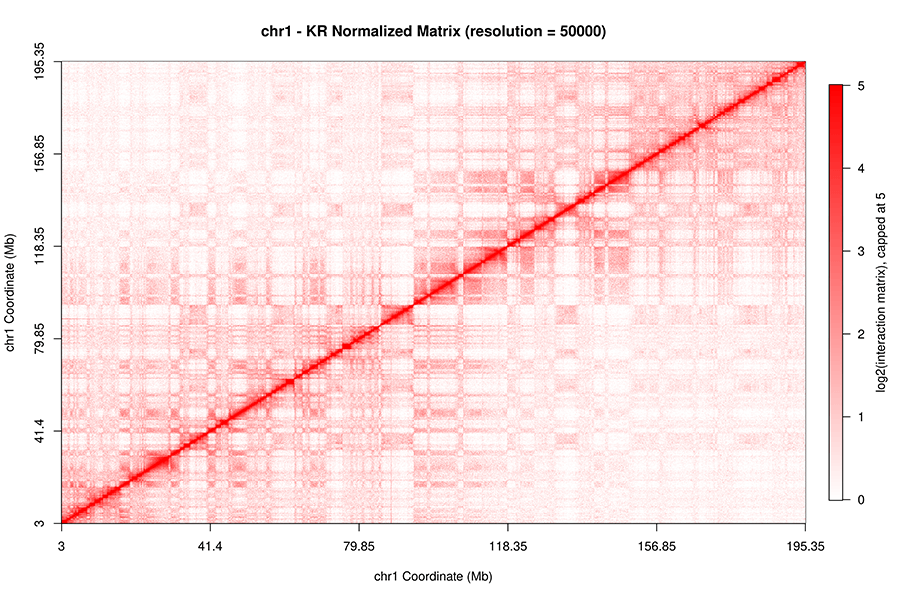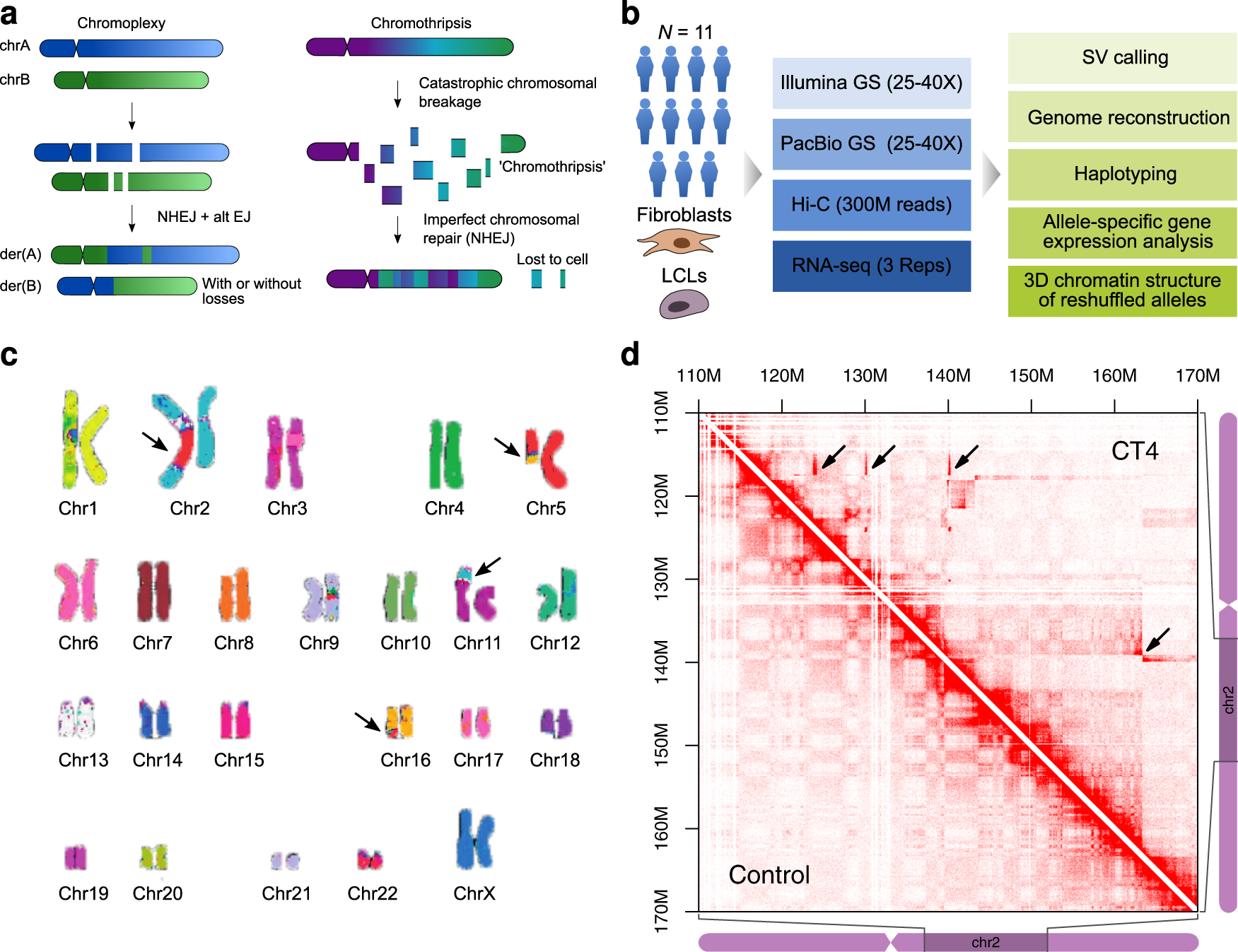
Integration of Hi-C with short and long-read genome sequencing reveals the structure of germline rearranged genomes | Nature Communications

Liquid chromatin Hi-C characterizes compartment-dependent chromatin interaction dynamics | Nature Genetics

Hi-C 2.0: An Optimized Hi-C Procedure for High-Resolution Genome-Wide Mapping of Chromosome Conformation | bioRxiv
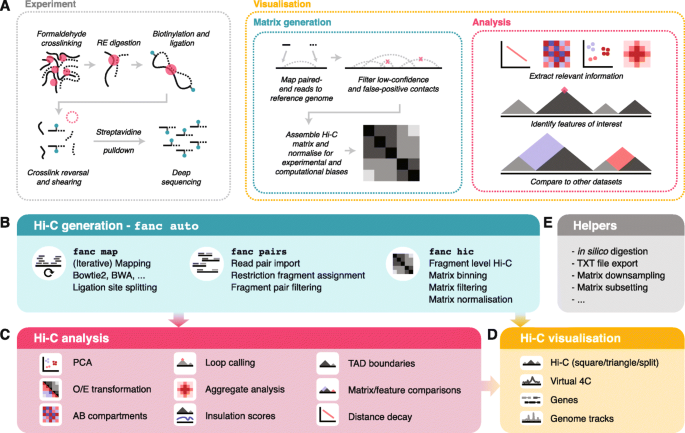
FAN-C: a feature-rich framework for the analysis and visualisation of chromosome conformation capture data | Genome Biology | Full Text

Hi-C 2.0: An optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation - ScienceDirect

BL-Hi-C is an efficient and sensitive approach for capturing structural and regulatory chromatin interactions | Nature Communications

Hi‐C 3.0: Improved Protocol for Genome‐Wide Chromosome Conformation Capture - Lafontaine - 2021 - Current Protocols - Wiley Online Library
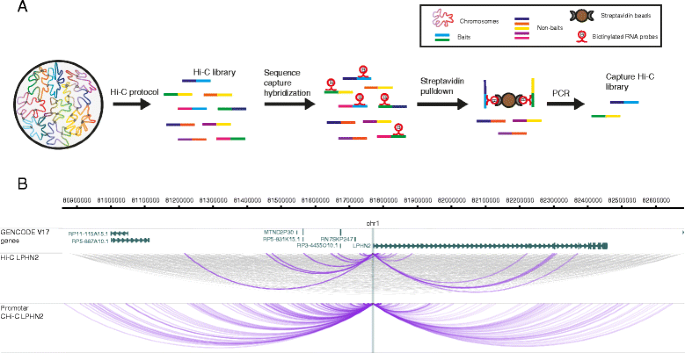
CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data | Genome Biology | Full Text