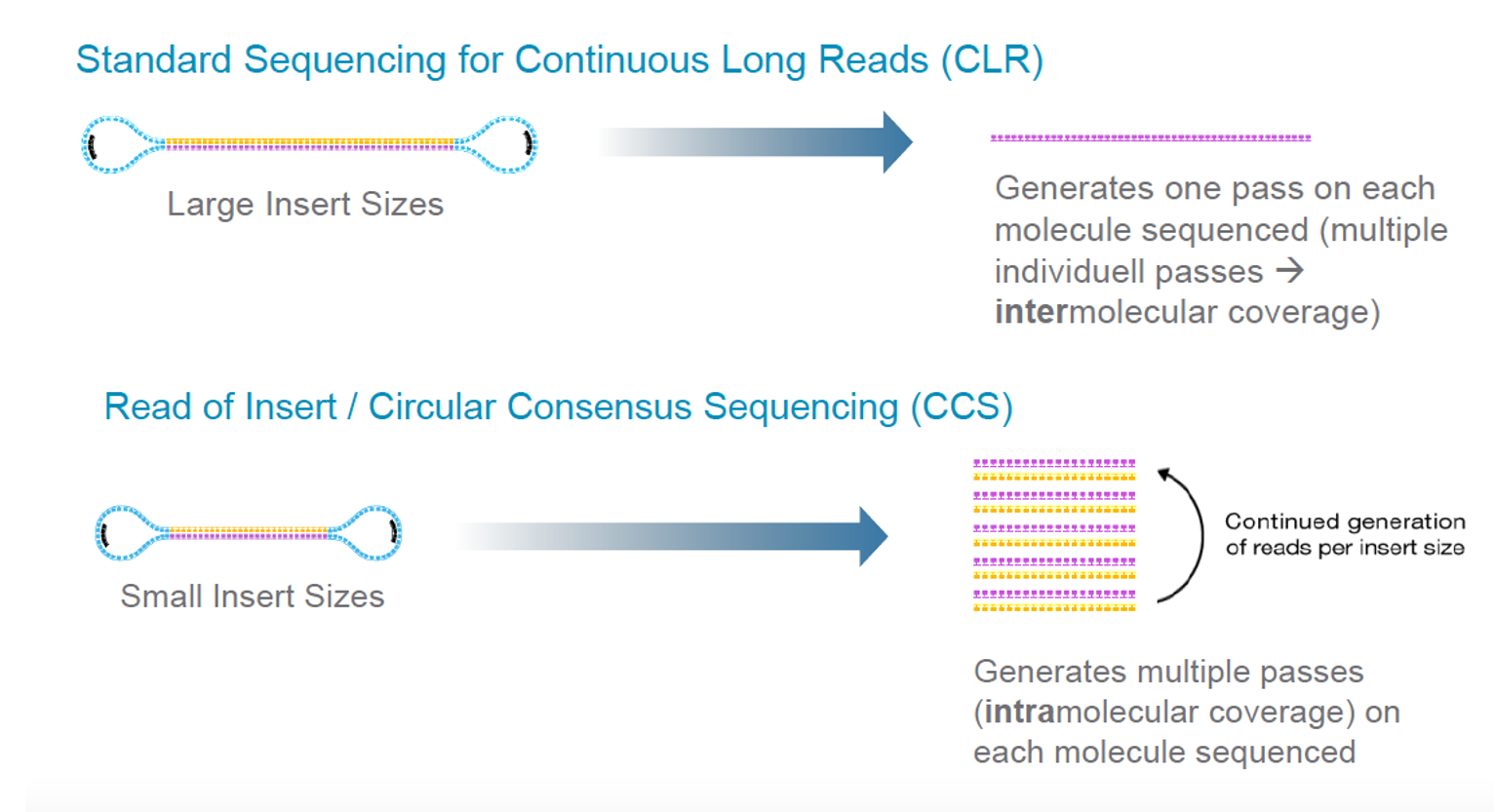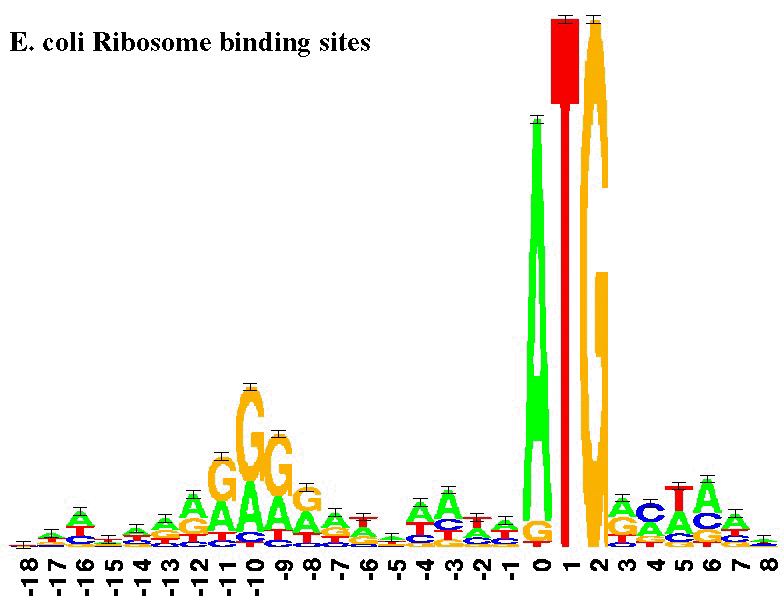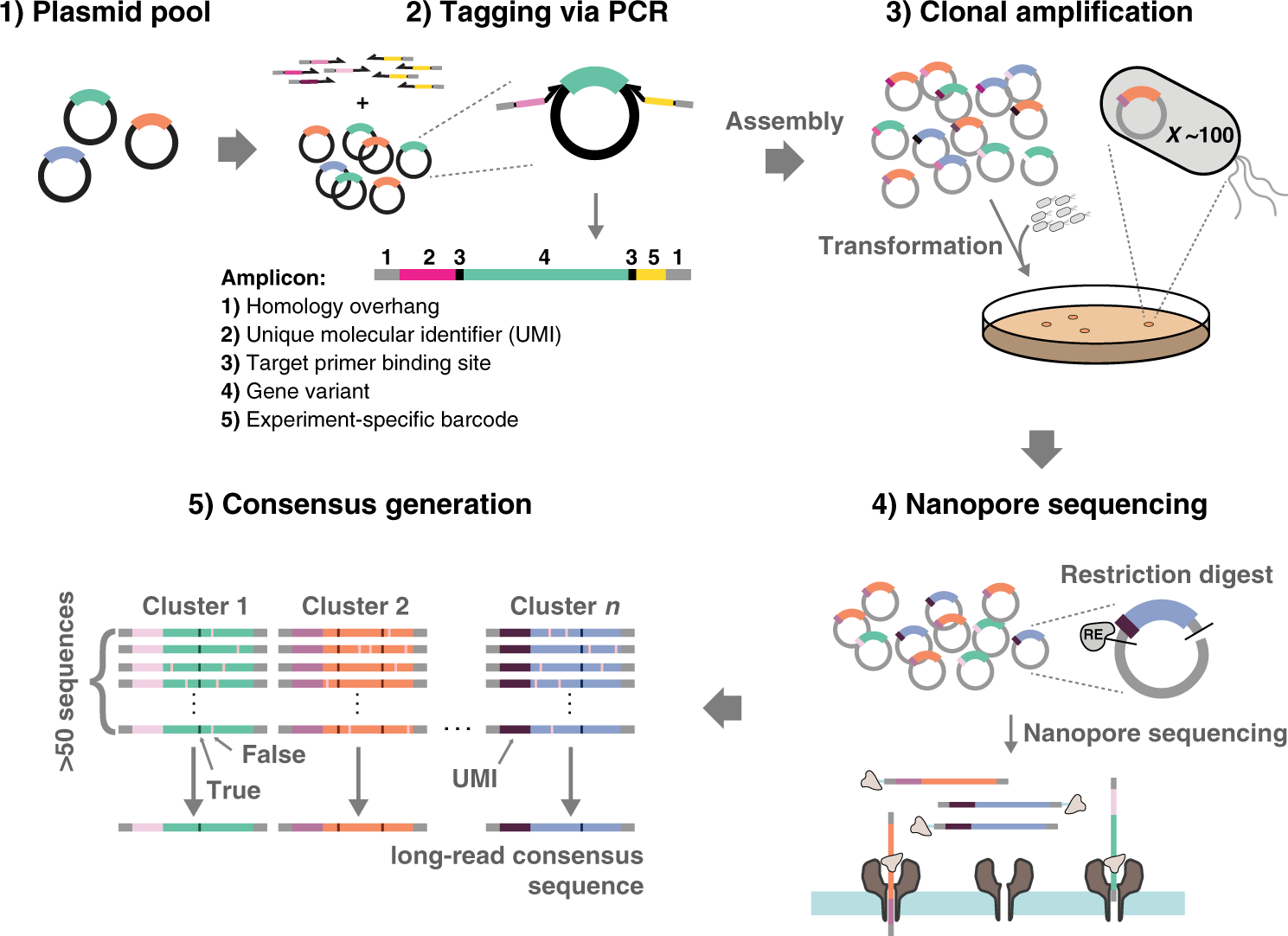
UMI-linked consensus sequencing enables phylogenetic analysis of directed evolution | Nature Communications

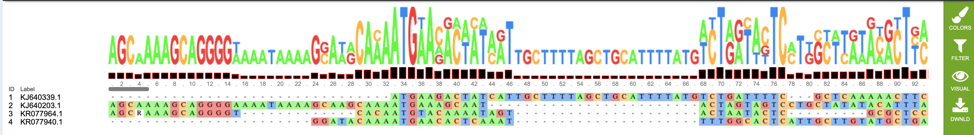
A) The number of consensus sequences generated by the pipelines that... | Download Scientific Diagram
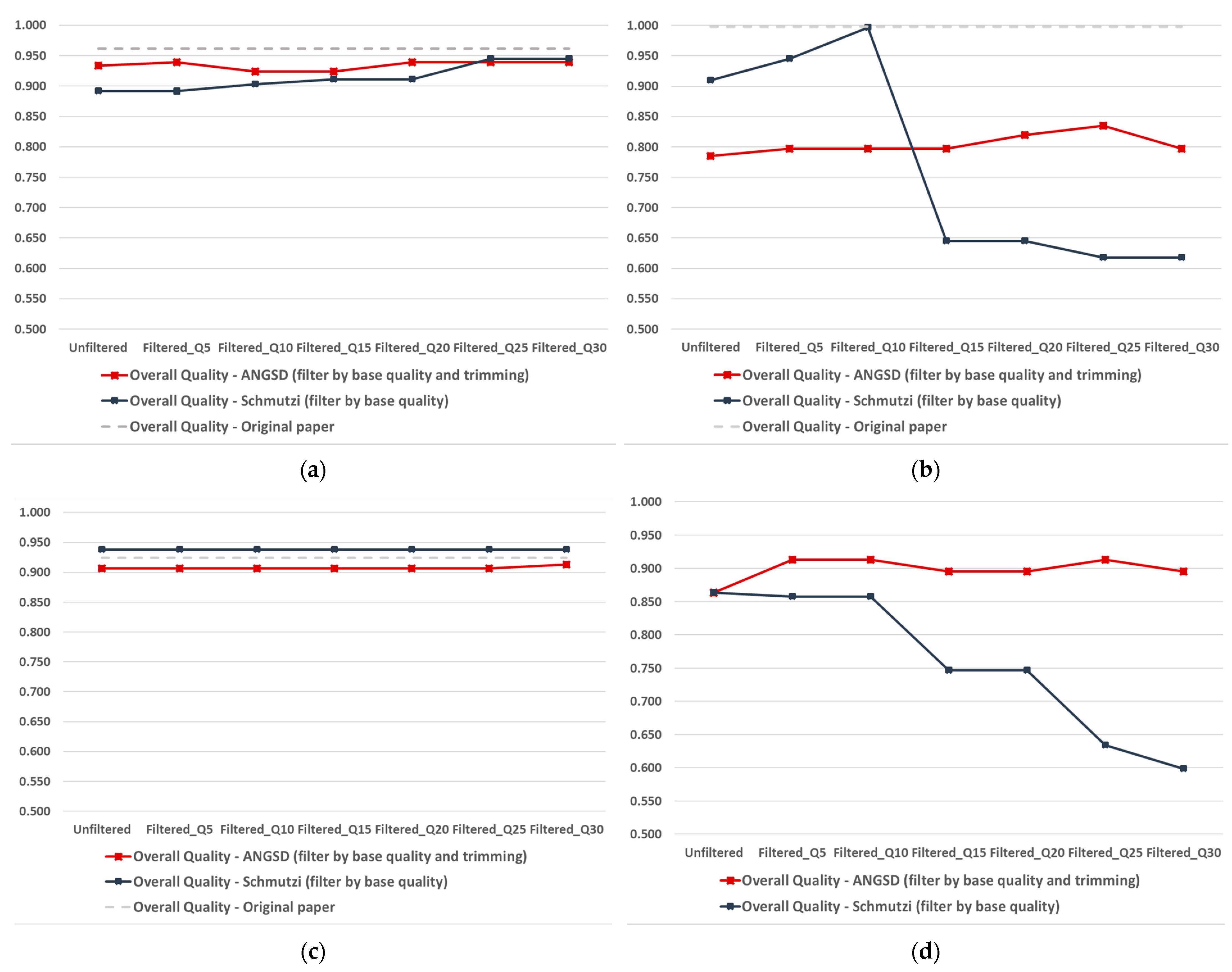
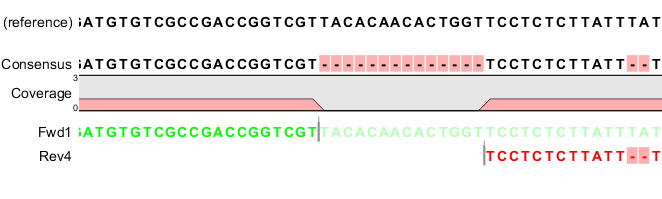
IJMS | Free Full-Text | Mitochondrial DNA Consensus Calling and Quality Filtering for Constructing Ancient Human Mitogenomes: Comparison of Two Widely Applied Methods

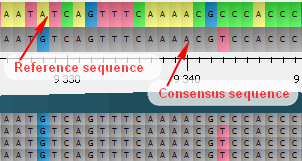
Building a consensus sequence with vcf files | by August Guang | Computational Biology Core | Medium




![PipeCoV: a pipeline for SARS-CoV-2 genome assembly, annotation and variant identification [PeerJ] PipeCoV: a pipeline for SARS-CoV-2 genome assembly, annotation and variant identification [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2022/13300/1/fig-4-2x.jpg)