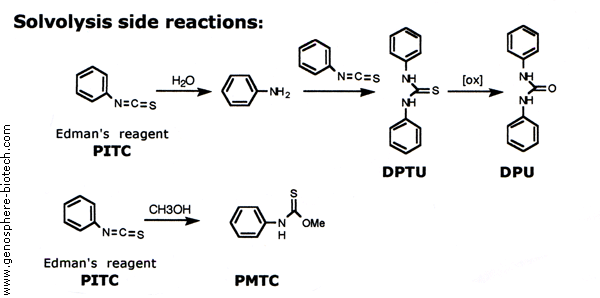
Determination of sequence and absolute configuration of peptide amino acids by HPLC–MS/CD-based detection of liberated N-terminus phenylthiohydantoin amino acids | Scientific Reports
Why Choose Creative Proteomics? Applications Methods of N-terminal Sequencing Key Features Sample Preparation Guideline You will
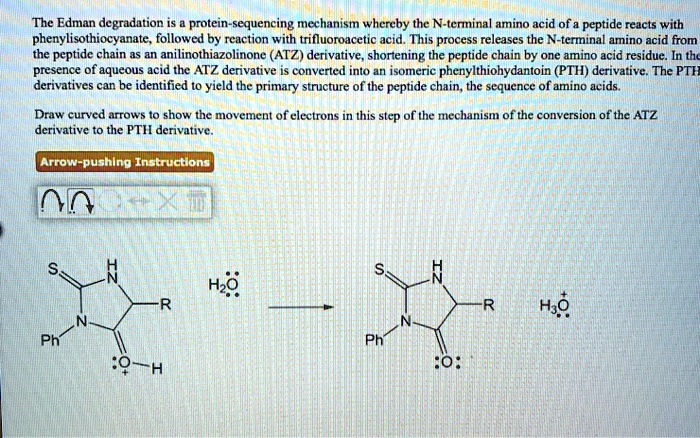
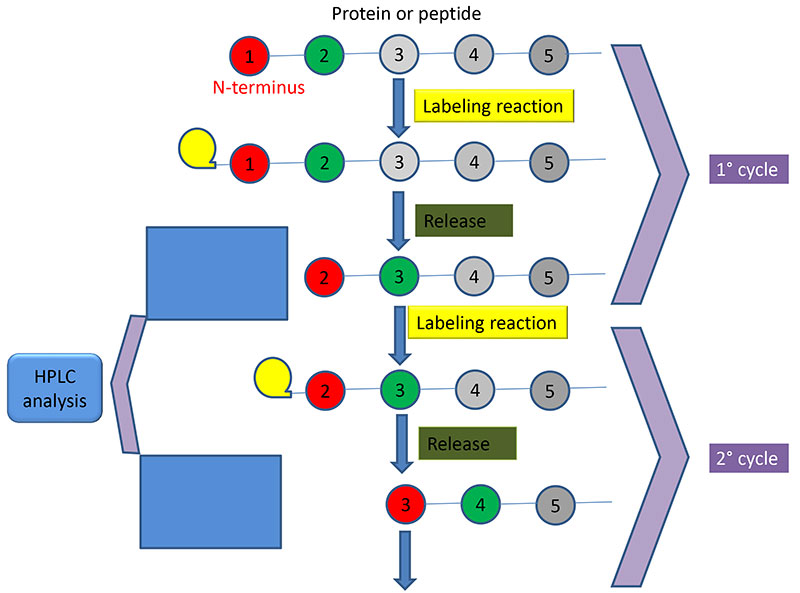
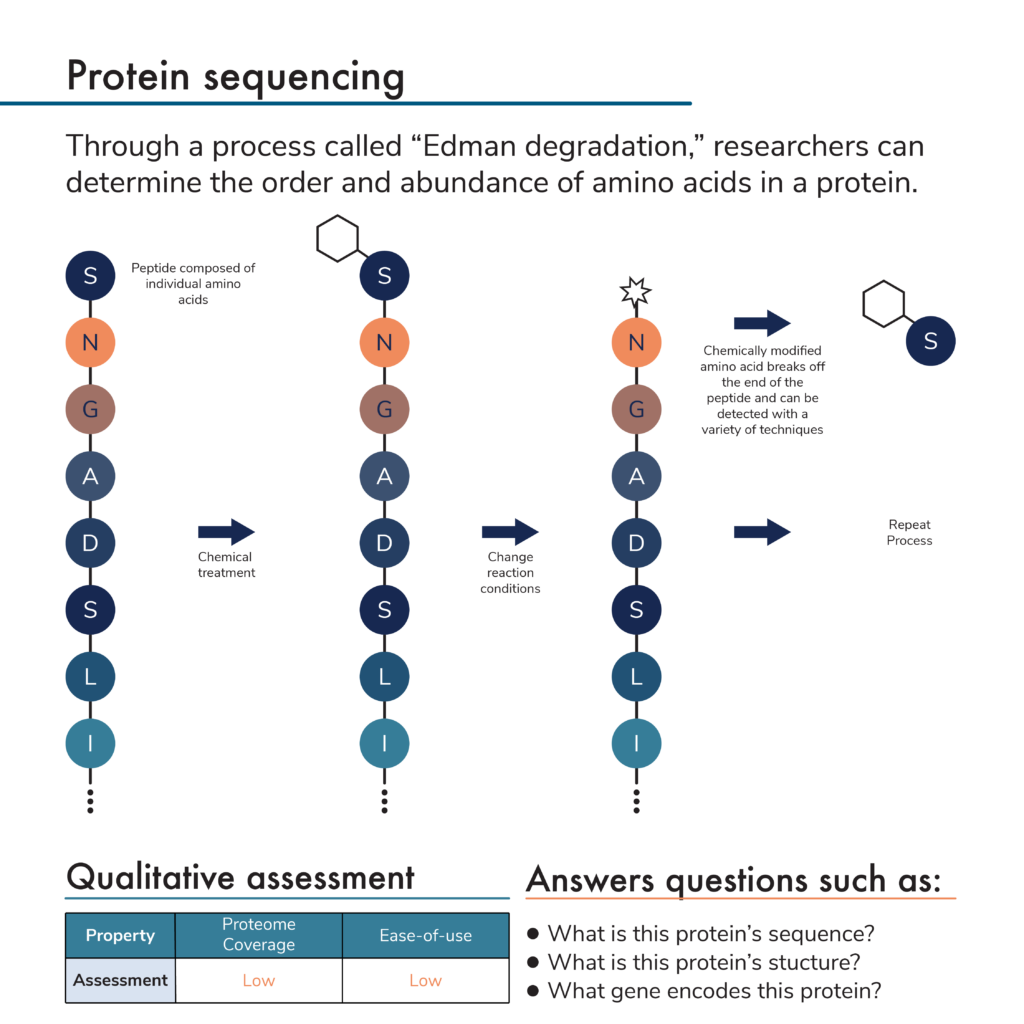
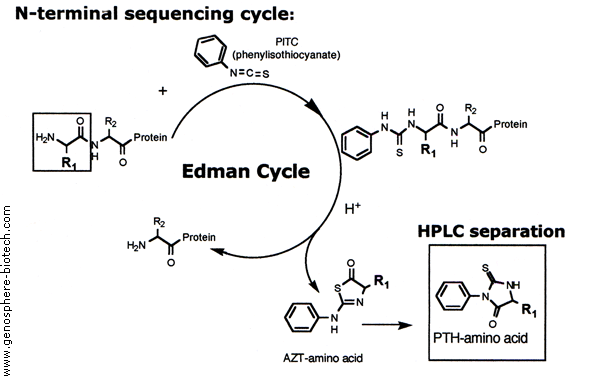
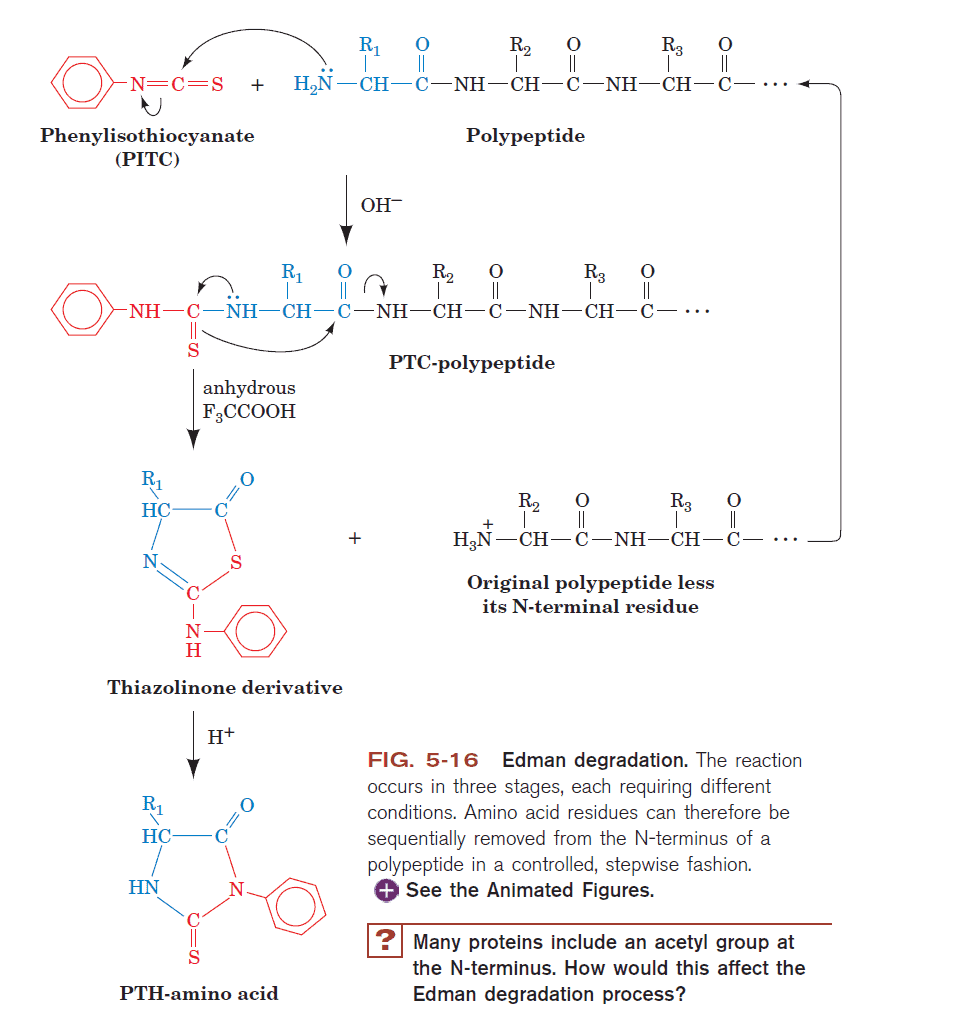
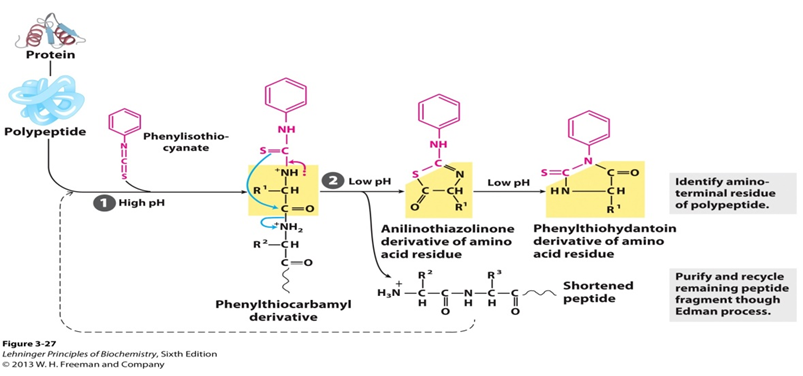
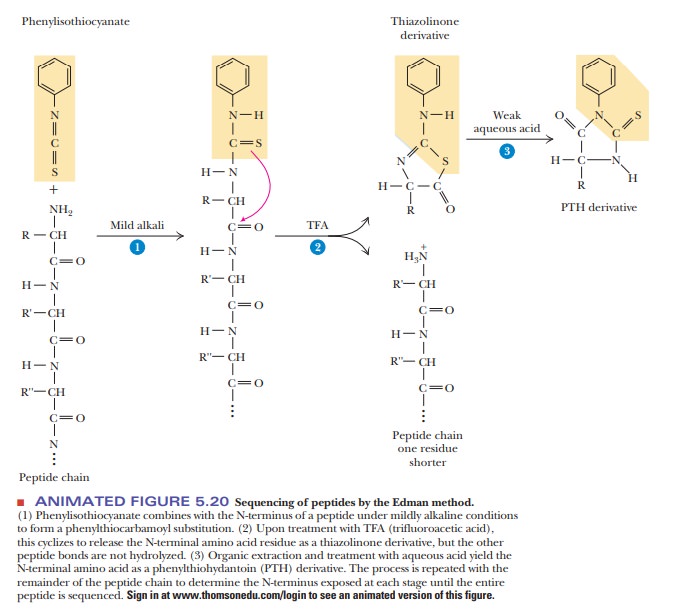
SOLVED: The Edman degradation protein-sequencing mechanism whereby the N-terminal amino acid of peptide reacts with phenylisothiocyanate , followed by reaction with trifluoroacetic acid. This process releases the N-terminal amino acid from the