Predicting Shine–Dalgarno Sequence Locations Exposes Genome Annotation Errors | PLOS Computational Biology

Influences on gene expression in vivo by a Shine–Dalgarno sequence - Jin - 2006 - Molecular Microbiology - Wiley Online Library

The Highly Efficient Translation Initiation Region from the Escherichia coli rpsA Gene Lacks a Shine-Dalgarno Element | Journal of Bacteriology
![PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/ca6e3ce301334ffc5d3a7df6b8a8e5d2bc19ed16/56-Table2.1-1.png)
PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar
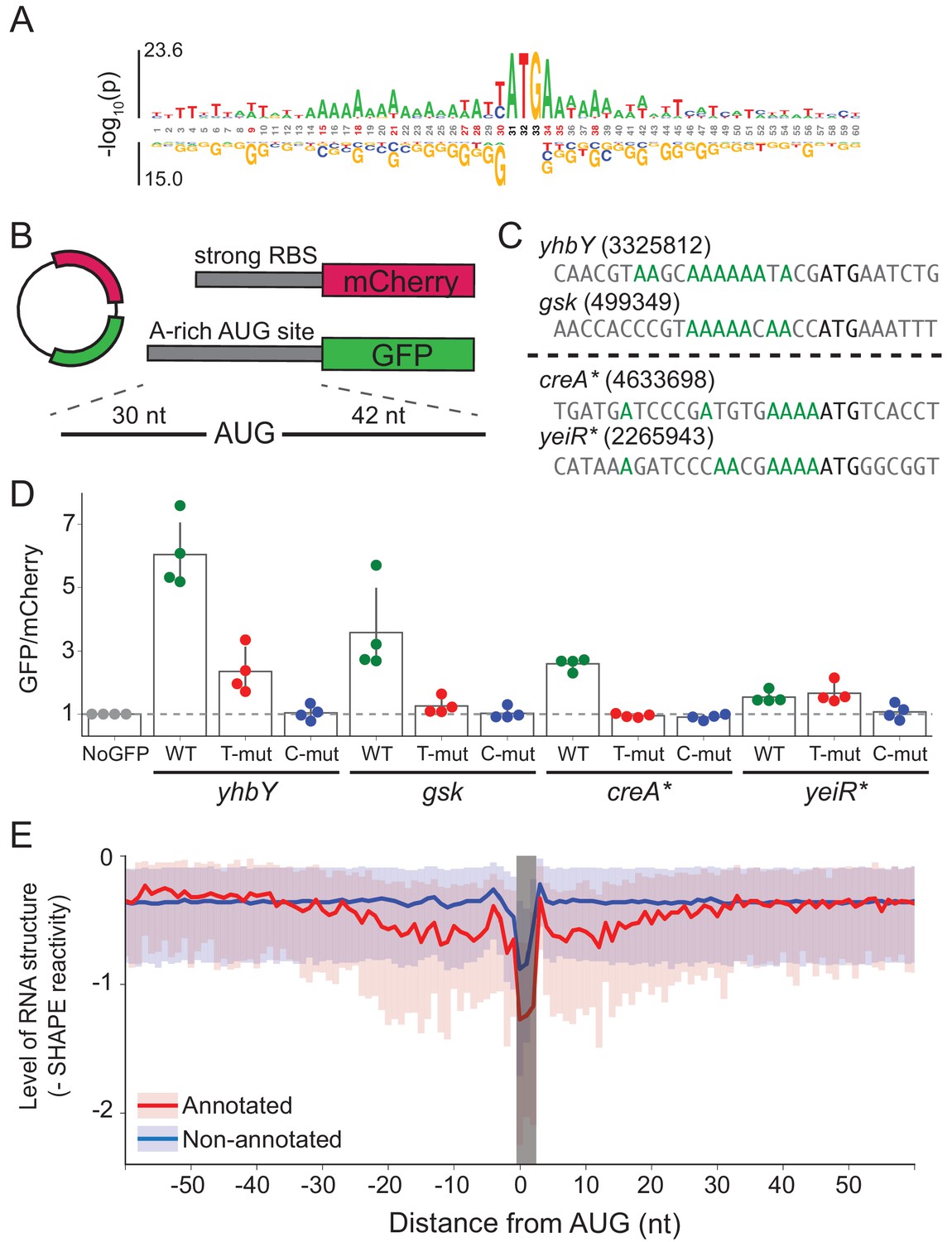
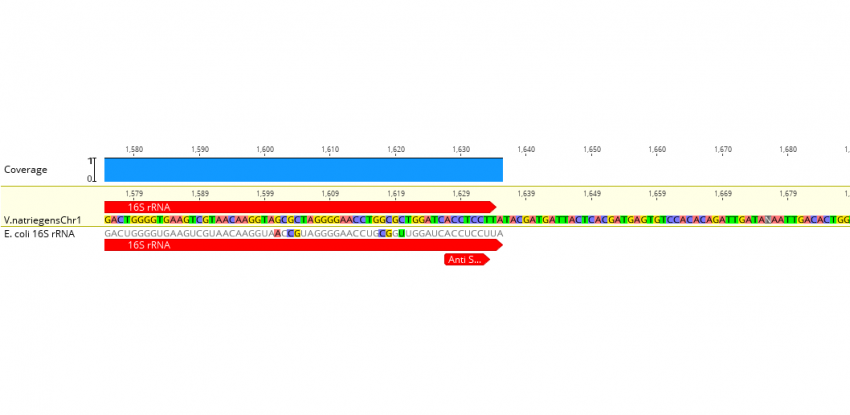
Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing | eLife
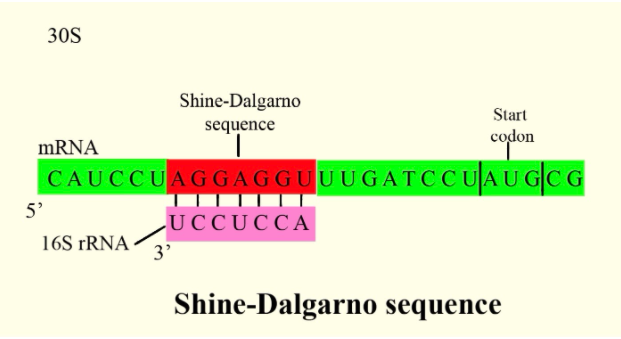
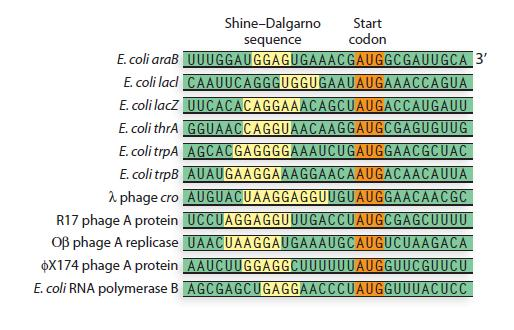
In prokaryotes, the ribosomes binding site on mRNA is called(a)Hogness sequence(b)Shine- Dalgarno sequence(c)Prinbow sequence(d)TATA- box

Leveraging genome-wide datasets to quantify the functional role of the anti- Shine–Dalgarno sequence in regulating translation efficiency | Open Biology

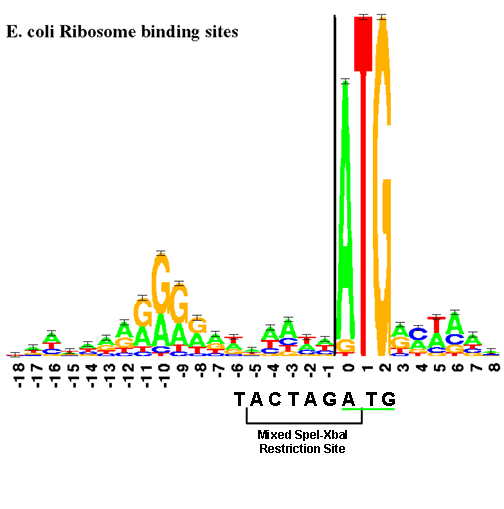
Sequence Logo for Shine-Dalgarno sequence in Escherichia coli. Sequence... | Download Scientific Diagram

Influence of the spacer region between the Shine–Dalgarno box and the start codon for fine‐tuning of the translation efficiency in Escherichia coli - Komarova - 2020 - Microbial Biotechnology - Wiley Online Library

Defining the anti-Shine-Dalgarno sequence interaction and quantifying its functional role in regulating translation efficiency | bioRxiv

Shine-Dalgarno sequence motifs (left) and Pribnow box (-10 box) motifs... | Download Scientific Diagram

Silent mutations in the Shine-Dalgarno-like sequence and the predicted... | Download Scientific Diagram

Figure 3 from Can The Insertion Of An E. coli Shine-Dalgarno Sequence Upstream Of M. ruber proA Of The proBA Operon Enhance Its Expression, As Measured By A Complementation Assay Using E.
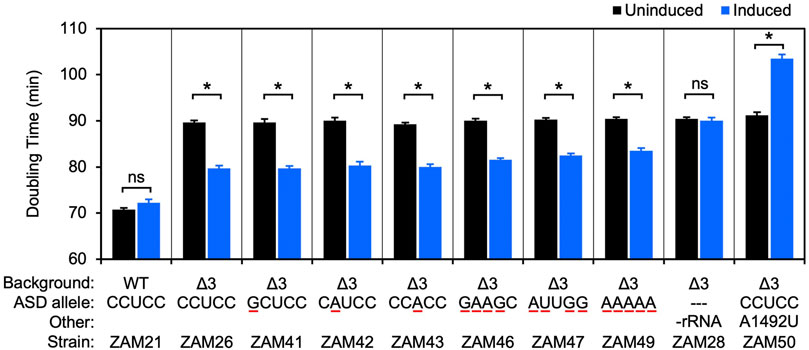
Frontiers | Comparative Analysis of anti-Shine- Dalgarno Function in Flavobacterium johnsoniae and Escherichia coli
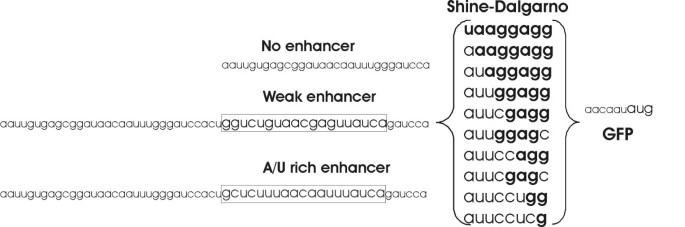
Translation initiation region sequence preferences in Escherichia coli | BMC Molecular Biology | Full Text

Extending the Spacing between the Shine–Dalgarno Sequence and P-Site Codon Reduces the Rate of mRNA Translocation - ScienceDirect

Accessibility of the Shine-Dalgarno Sequence Dictates N-Terminal Codon Bias in E. coli - ScienceDirect




![Solved 2. [ 2 points] This problem concerns the dsDNA gene | Chegg.com Solved 2. [ 2 points] This problem concerns the dsDNA gene | Chegg.com](https://media.cheggcdn.com/media/99e/99e1018b-6b24-4cd1-ae58-b4bfcdebd844/phpbavxuC)