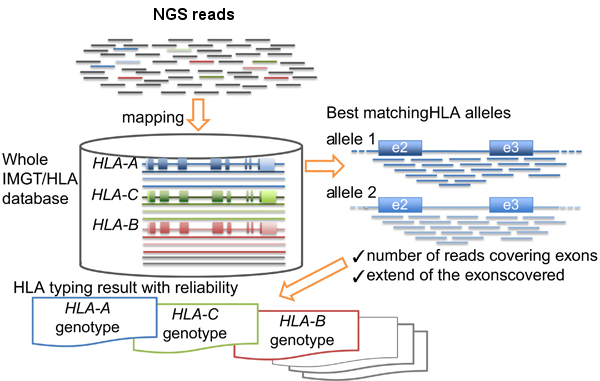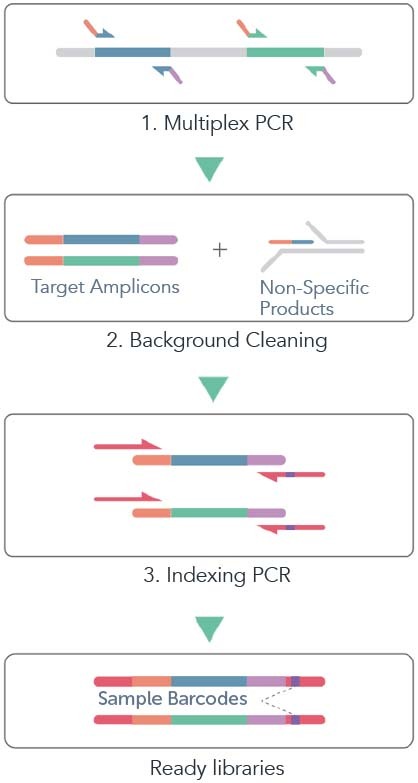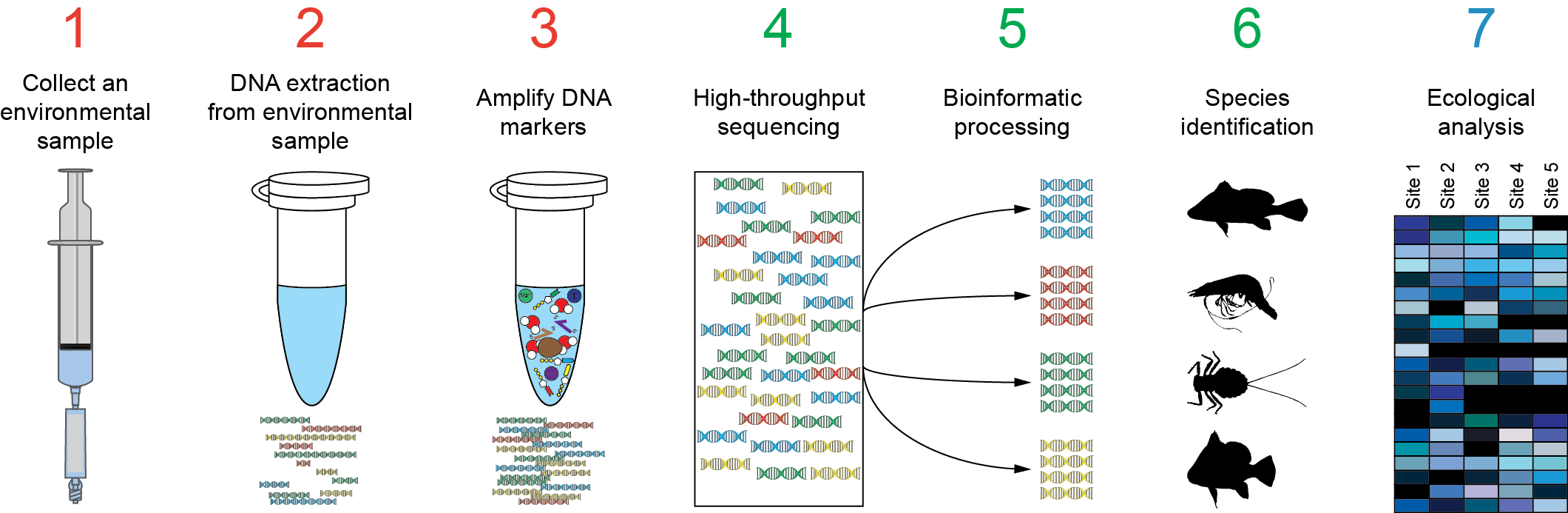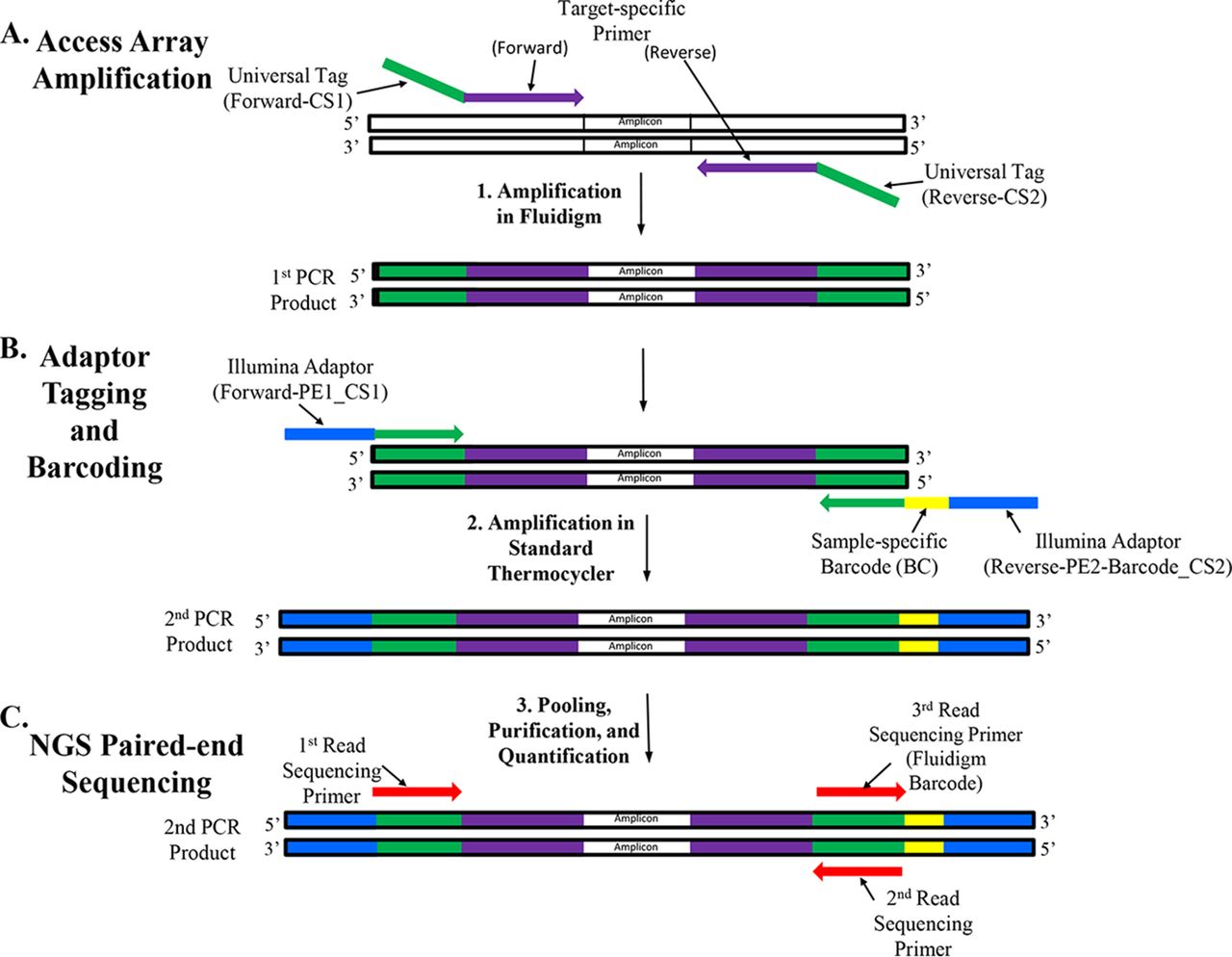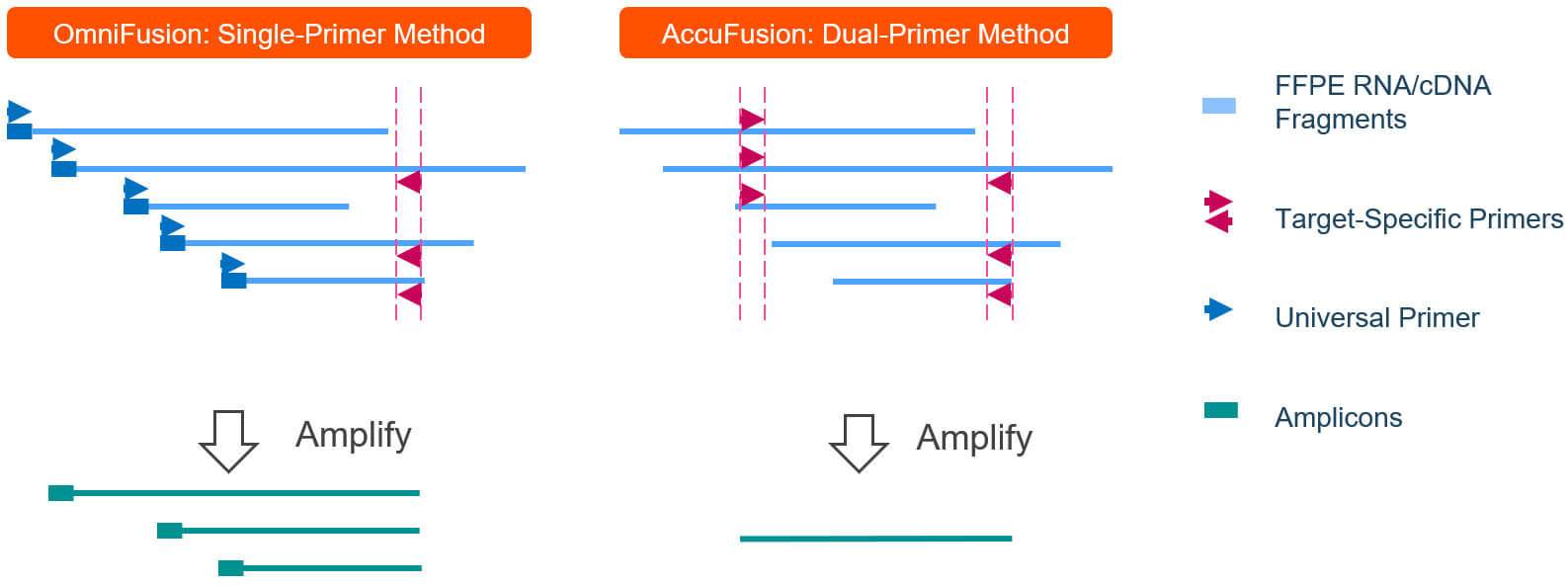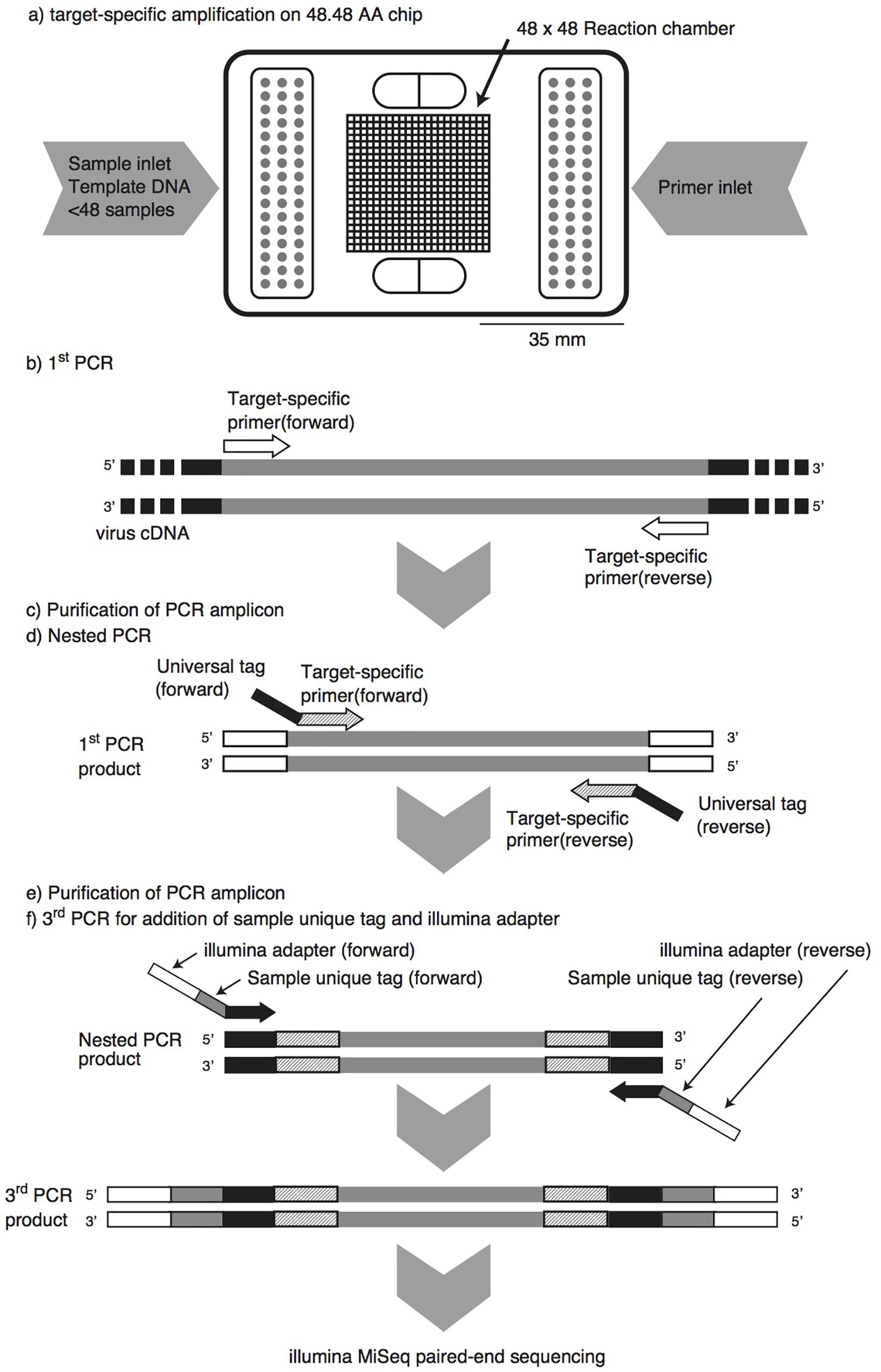
Frontiers | Microfluidic PCR Amplification and MiSeq Amplicon Sequencing Techniques for High-Throughput Detection and Genotyping of Human Pathogenic RNA Viruses in Human Feces, Sewage, and Oysters
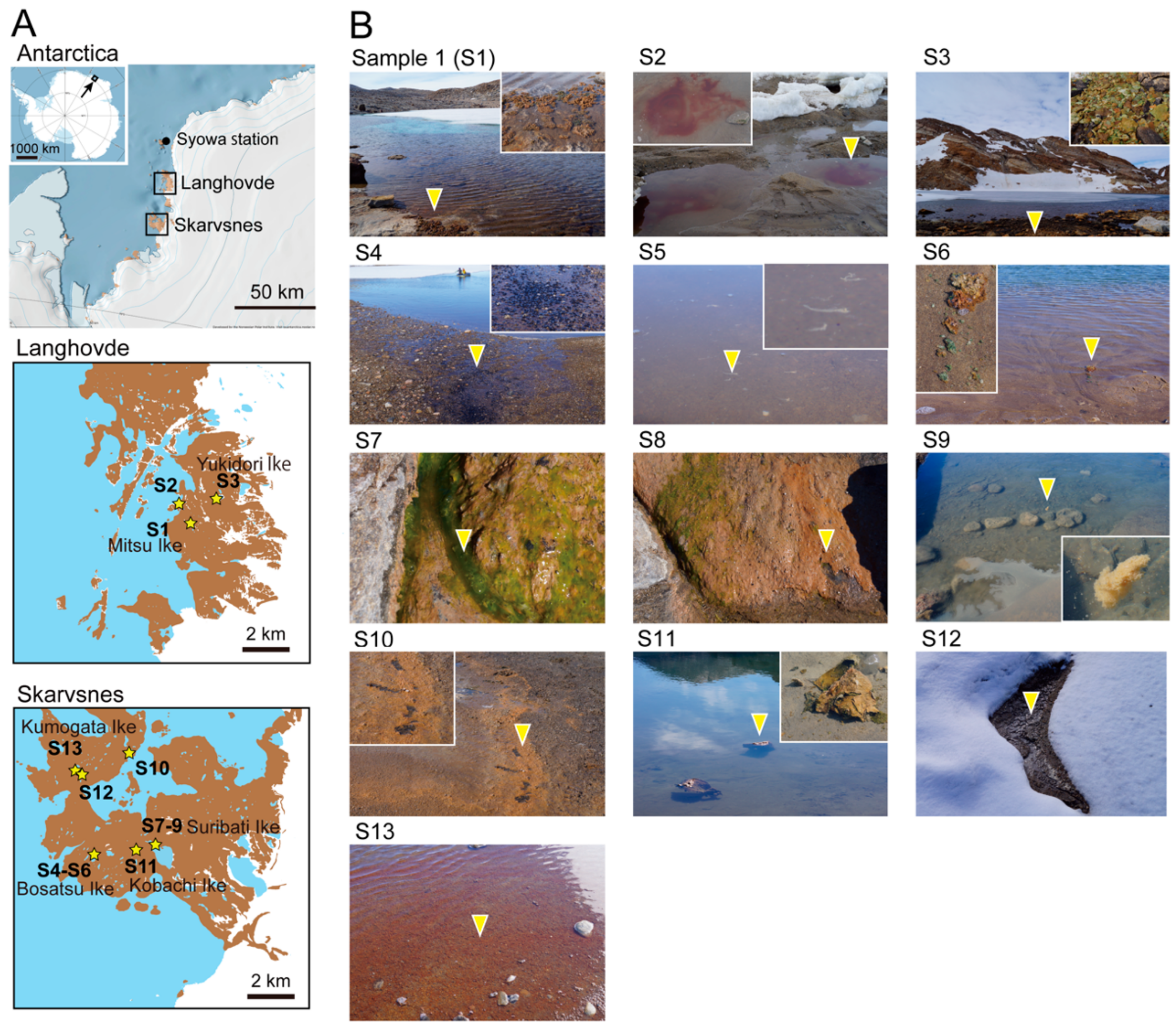
Microorganisms | Free Full-Text | Investigating Algal Communities in Lacustrine and Hydro-Terrestrial Environments of East Antarctica Using Deep Amplicon Sequencing

Keith Slotkin on Twitter: "Please vote for my lab's entry into the @BioRender figure competition: Ultra-deep DNA methylation patterns via amplicon sequencing. By postdoc Andrea McCue. https://t.co/CiKXWX0Cug https://t.co/PywCf3EY2Y" / Twitter

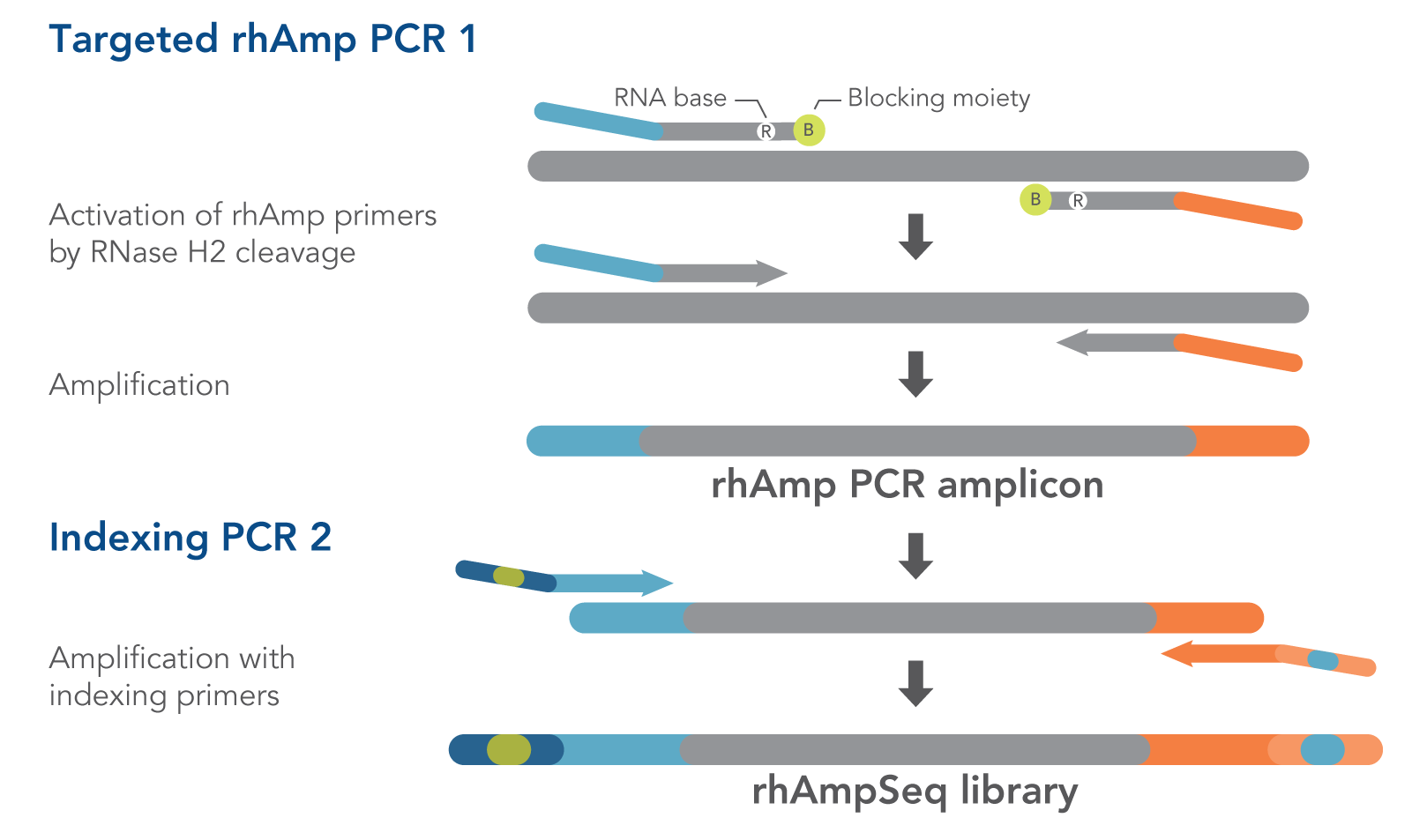
Ultrasensitive amplicon barcoding for next-generation sequencing facilitating sequence error and amplification-bias correction | Scientific Reports
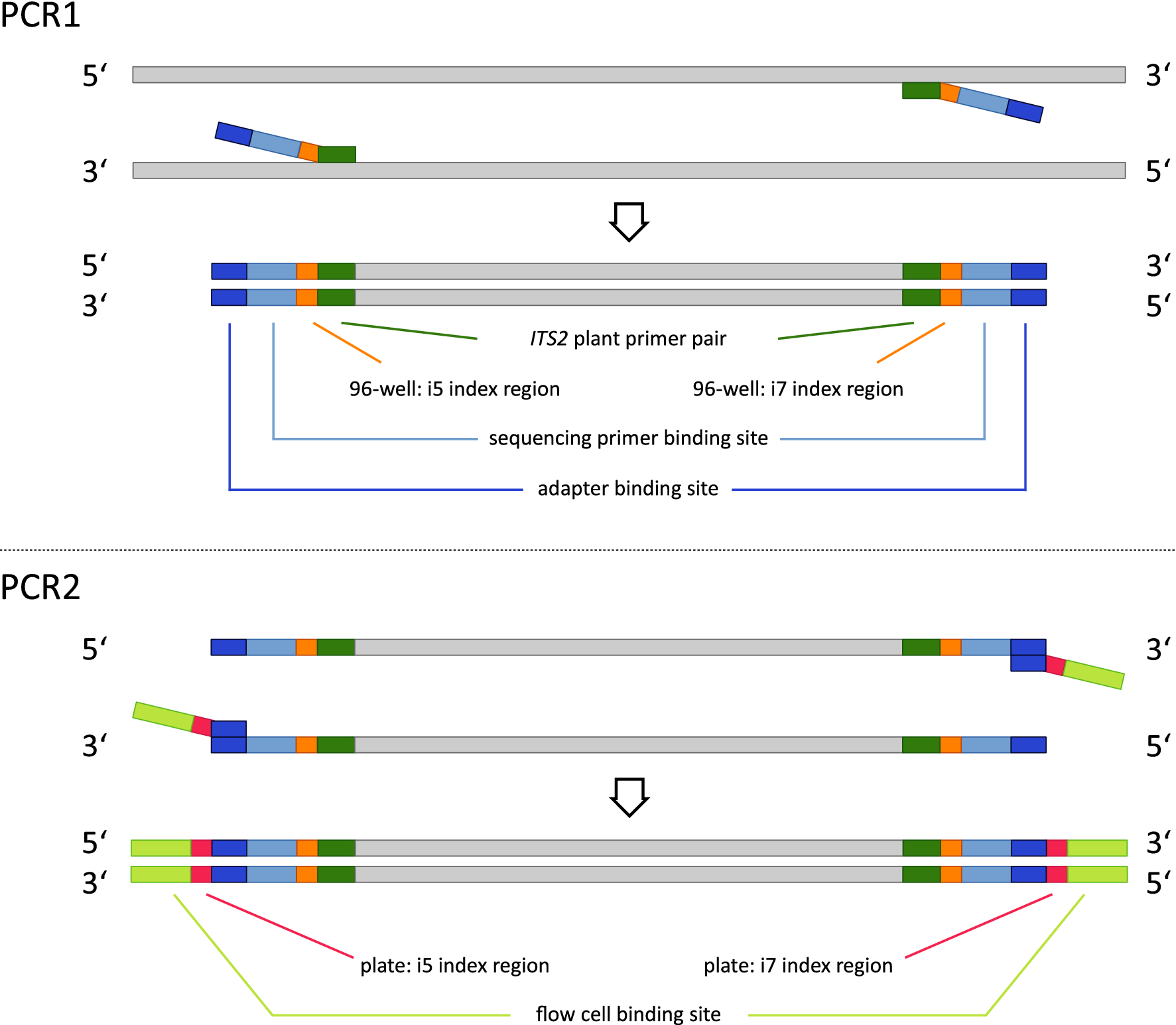
Handling of targeted amplicon sequencing data focusing on index hopping and demultiplexing using a nested metabarcoding approach in ecology | Scientific Reports
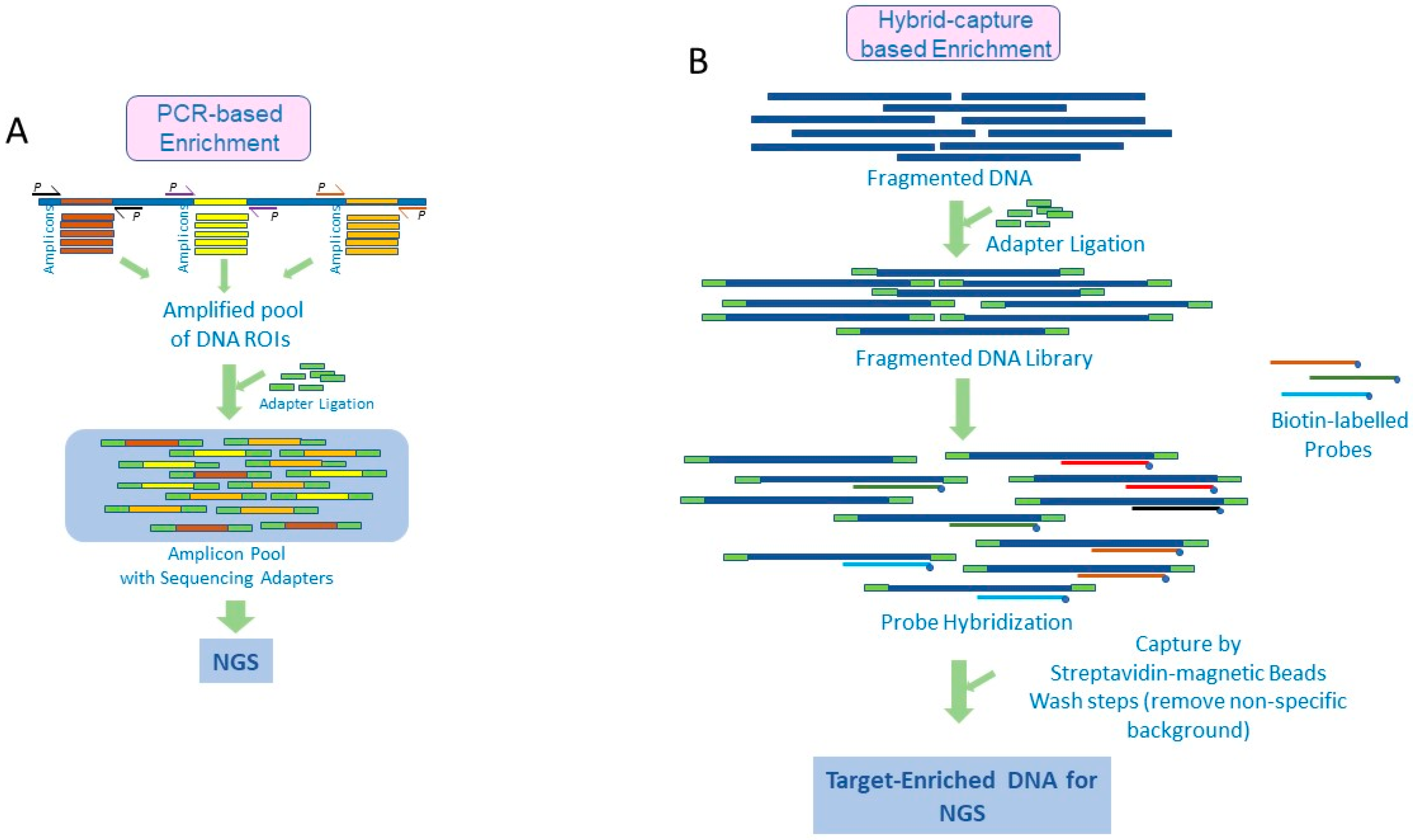
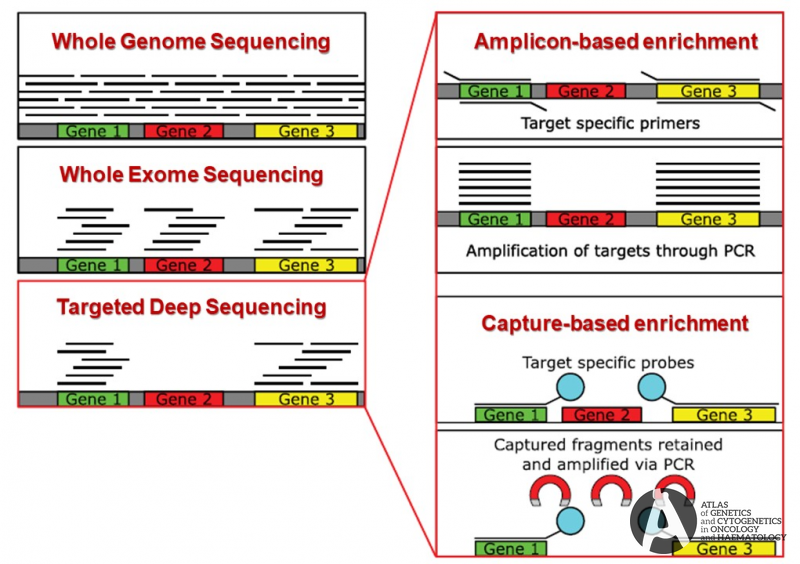
Diagnostics | Free Full-Text | Target Enrichment Approaches for Next-Generation Sequencing Applications in Oncology
Clonal Structure of ATLL Patients Analyzed by Amplicon Deep Sequencing... | Download Scientific Diagram