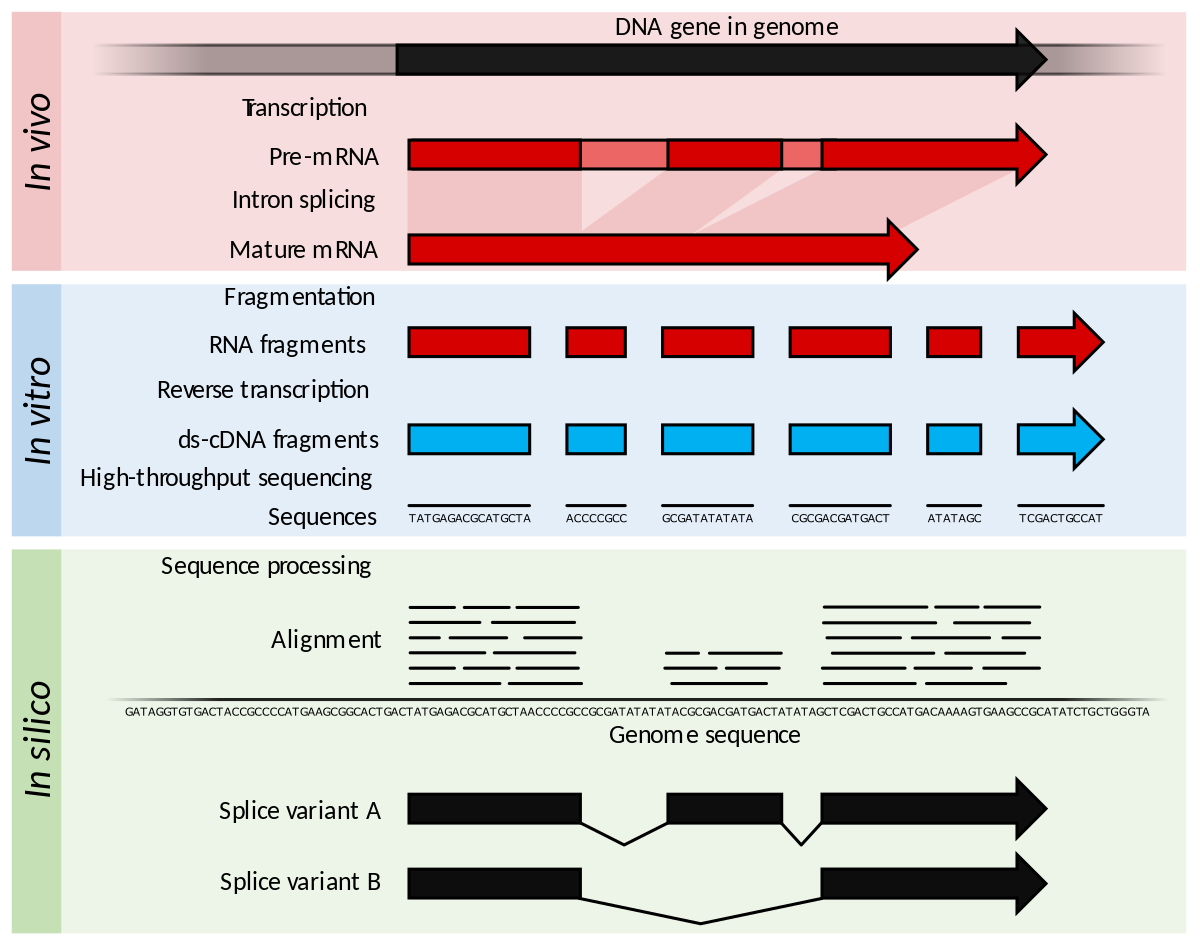
Transcriptomic Profiling Using Next Generation Sequencing - Advances, Advantages, and Challenges | IntechOpen
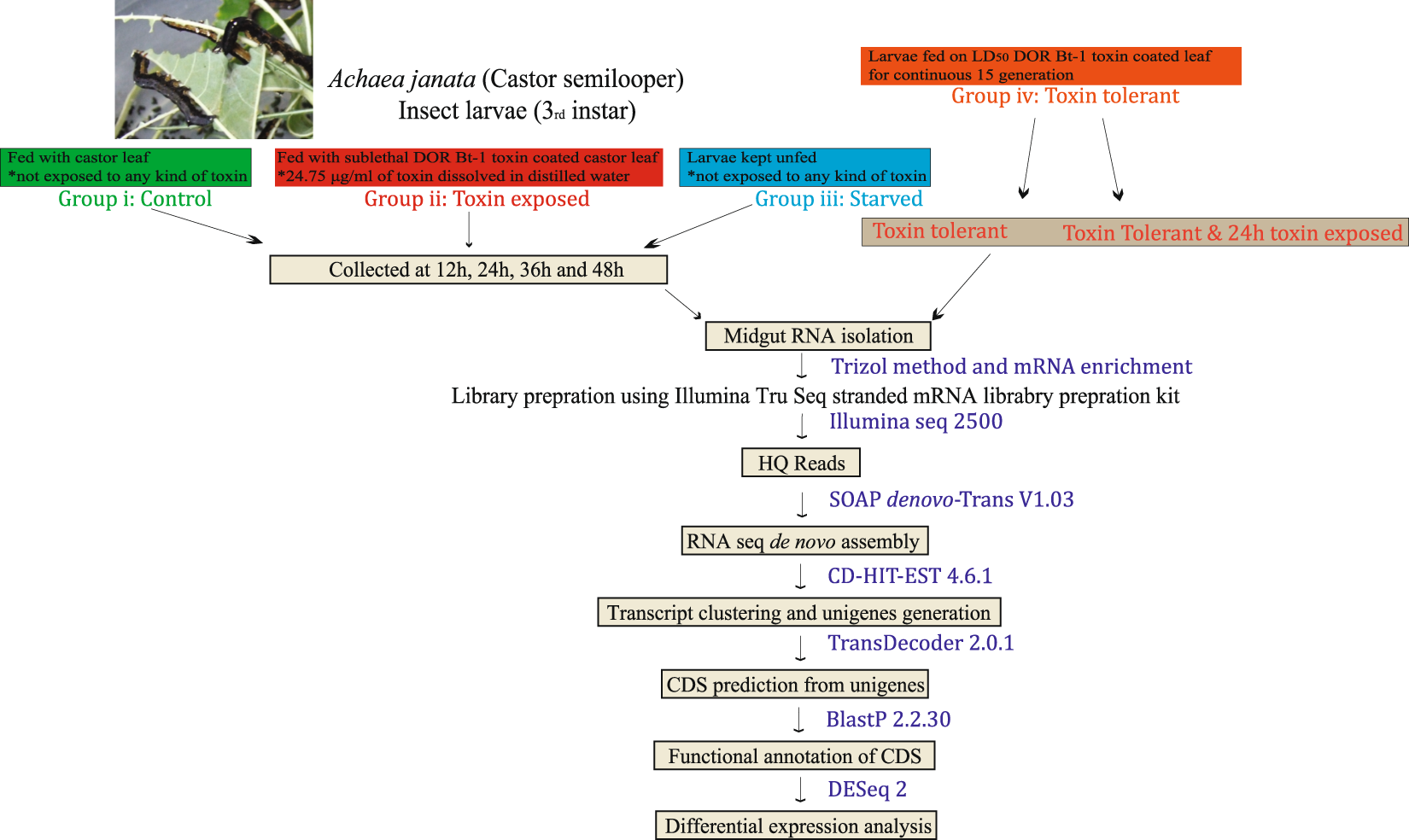
RNA-Seq analysis and de novo transcriptome assembly of Cry toxin susceptible and tolerant Achaea janata larvae | Scientific Data
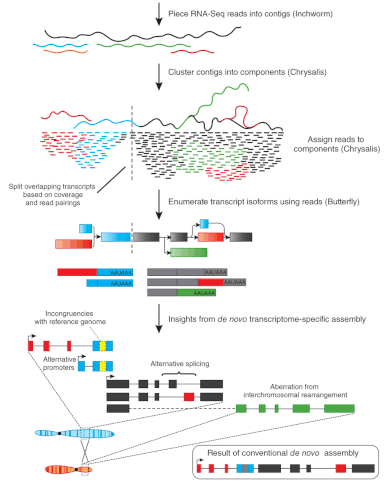
Bioinformatics challenges in de novo transcriptome assembly using short read sequences in the absence of a reference genome sequence - Natural Product Reports (RSC Publishing) DOI:10.1039/C3NP20099J

Transcriptomics RNASeq | Transcriptome Analysis – 1010Genome | Quality NGS Bioinformatics Data Analysis Services

PDF) Bioinformatics challenges in de novo transcriptome assembly using short read sequences in the absence of a reference genome sequence

A decade of de novo transcriptome assembly: Are we there yet? - Hölzer - 2021 - Molecular Ecology Resources - Wiley Online Library
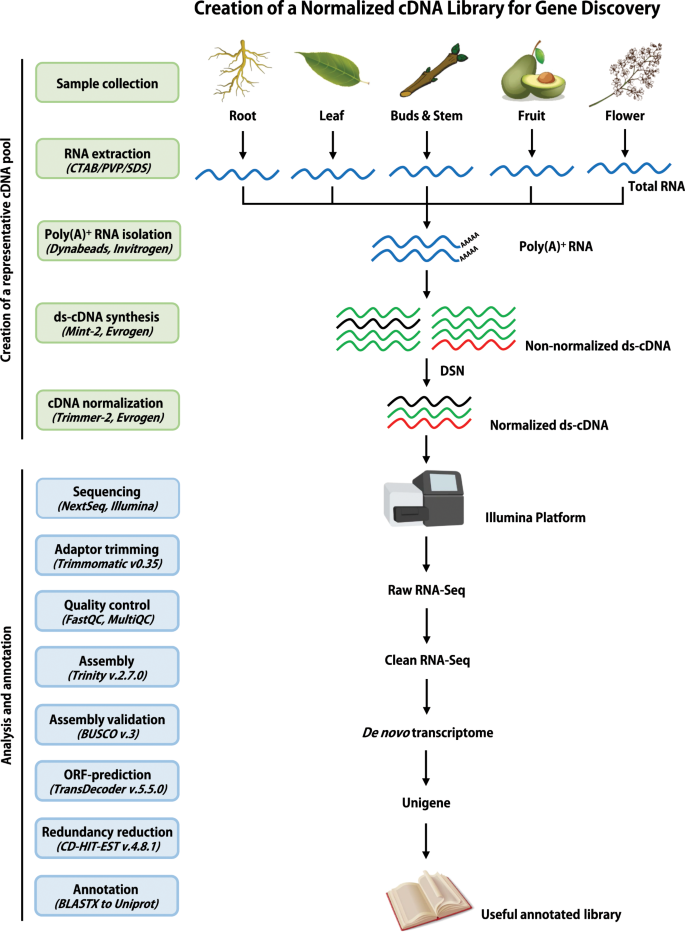
De novo transcriptome assembly and annotation for gene discovery in avocado, macadamia and mango | Scientific Data

TransPi – a comprehensive TRanscriptome ANalysiS PIpeline for de novo transcriptome assembly | RNA-Seq Blog
![PDF] Simultaneous Transcriptome Analysis of Sorghum and Bipolaris sorghicola by Using RNA-seq in Combination with De Novo Transcriptome Assembly | Semantic Scholar PDF] Simultaneous Transcriptome Analysis of Sorghum and Bipolaris sorghicola by Using RNA-seq in Combination with De Novo Transcriptome Assembly | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/9c859a8b24d8df7b46671c73221094087ababc81/3-Figure1-1.png)
PDF] Simultaneous Transcriptome Analysis of Sorghum and Bipolaris sorghicola by Using RNA-seq in Combination with De Novo Transcriptome Assembly | Semantic Scholar
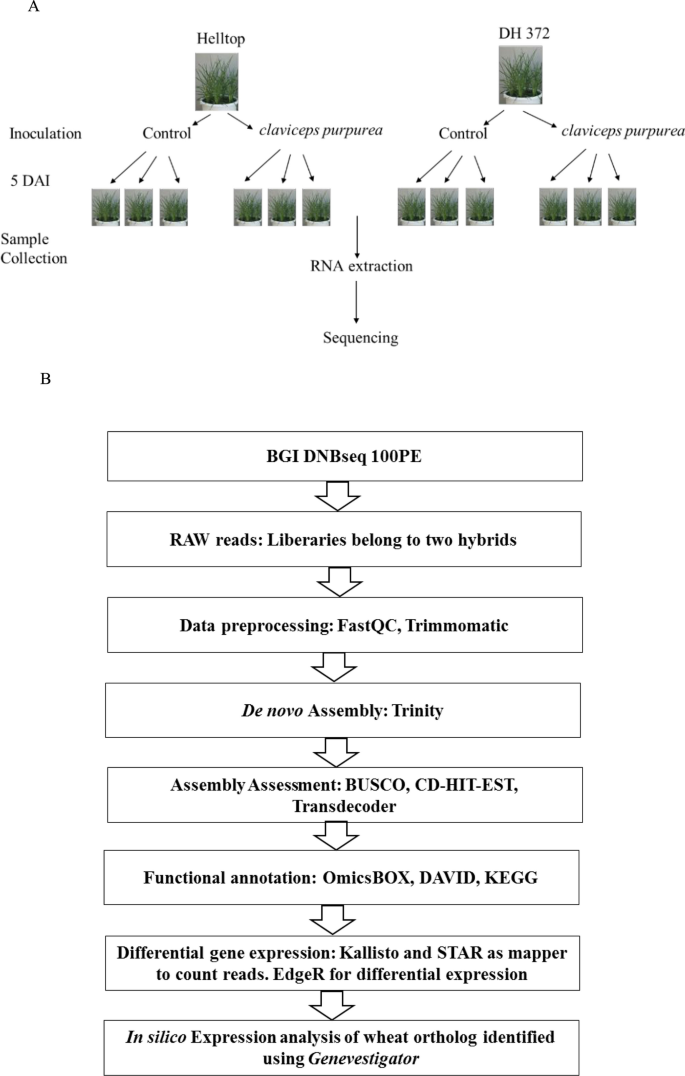
De novo transcriptome assembly, functional annotation, and expression profiling of rye (Secale cereale L.) hybrids inoculated with ergot (Claviceps purpurea) | Scientific Reports
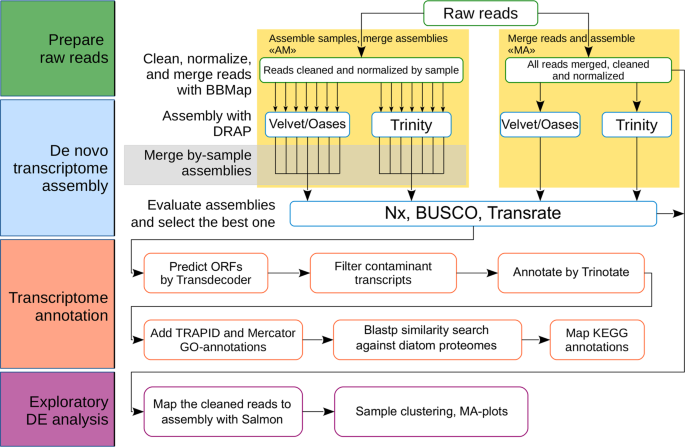
De novo transcriptome assembly and analysis of the freshwater araphid diatom Fragilaria radians, Lake Baikal | Scientific Data
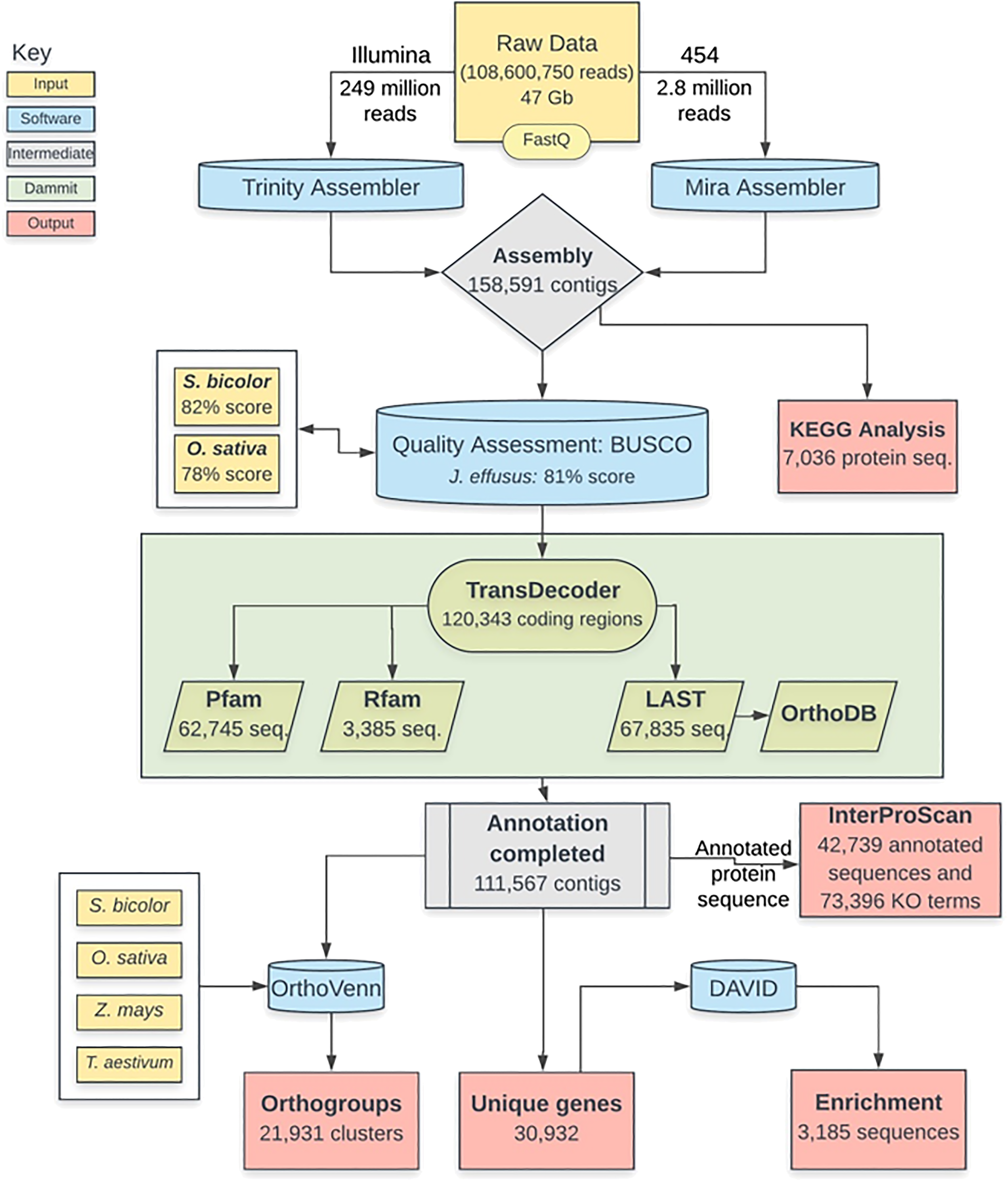
RNA-Seq analysis of soft rush (Juncus effusus): transcriptome sequencing, de novo assembly, annotation, and polymorphism identification | BMC Genomics | Full Text
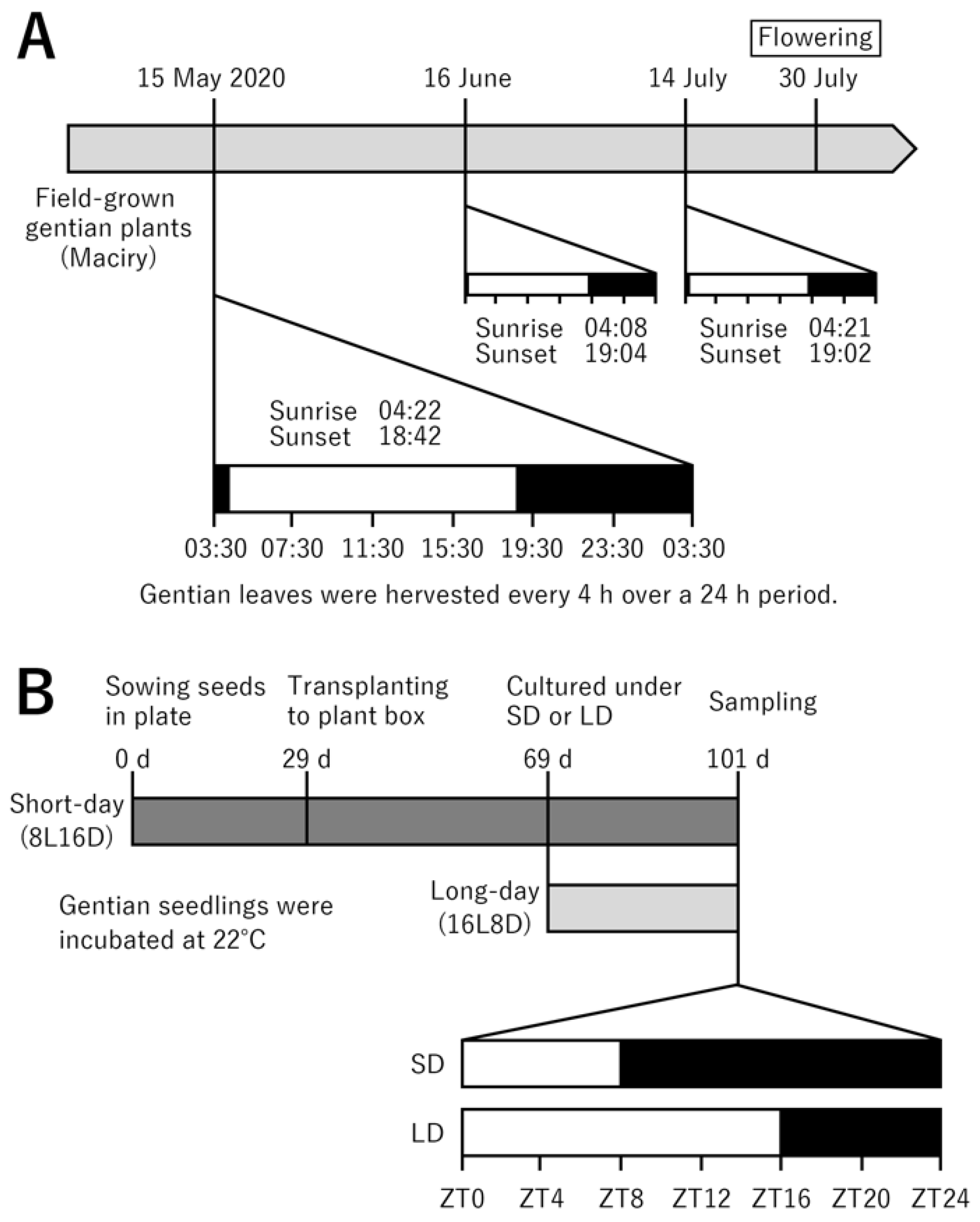
IJMS | Free Full-Text | De Novo Transcriptome Analysis Reveals Flowering-Related Genes That Potentially Contribute to Flowering-Time Control in the Japanese Cultivated Gentian Gentiana triflora
![PDF] Transcriptome Analysis for Non-Model Organism: Current Status and Best-Practices | Semantic Scholar PDF] Transcriptome Analysis for Non-Model Organism: Current Status and Best-Practices | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/3ff2d865c1d7ea30df8656c1961fa8a12395eb8b/3-Figure1-1.png)
PDF] Transcriptome Analysis for Non-Model Organism: Current Status and Best-Practices | Semantic Scholar

Bioinformatics challenges in de novo transcriptome assembly using short read sequences in the absence of a reference genome sequence. | Semantic Scholar

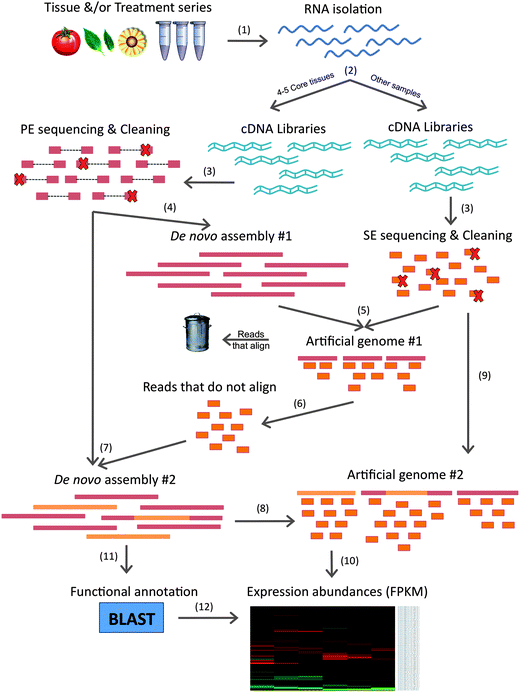
A Detailed Overview of the De Novo Transcriptome Assembly and Analysis... | Download Scientific Diagram

De novo Transcriptome Assembly & Differential Gene Expression Analysis | Quality NGS Bioinformatics Data Analysis Services