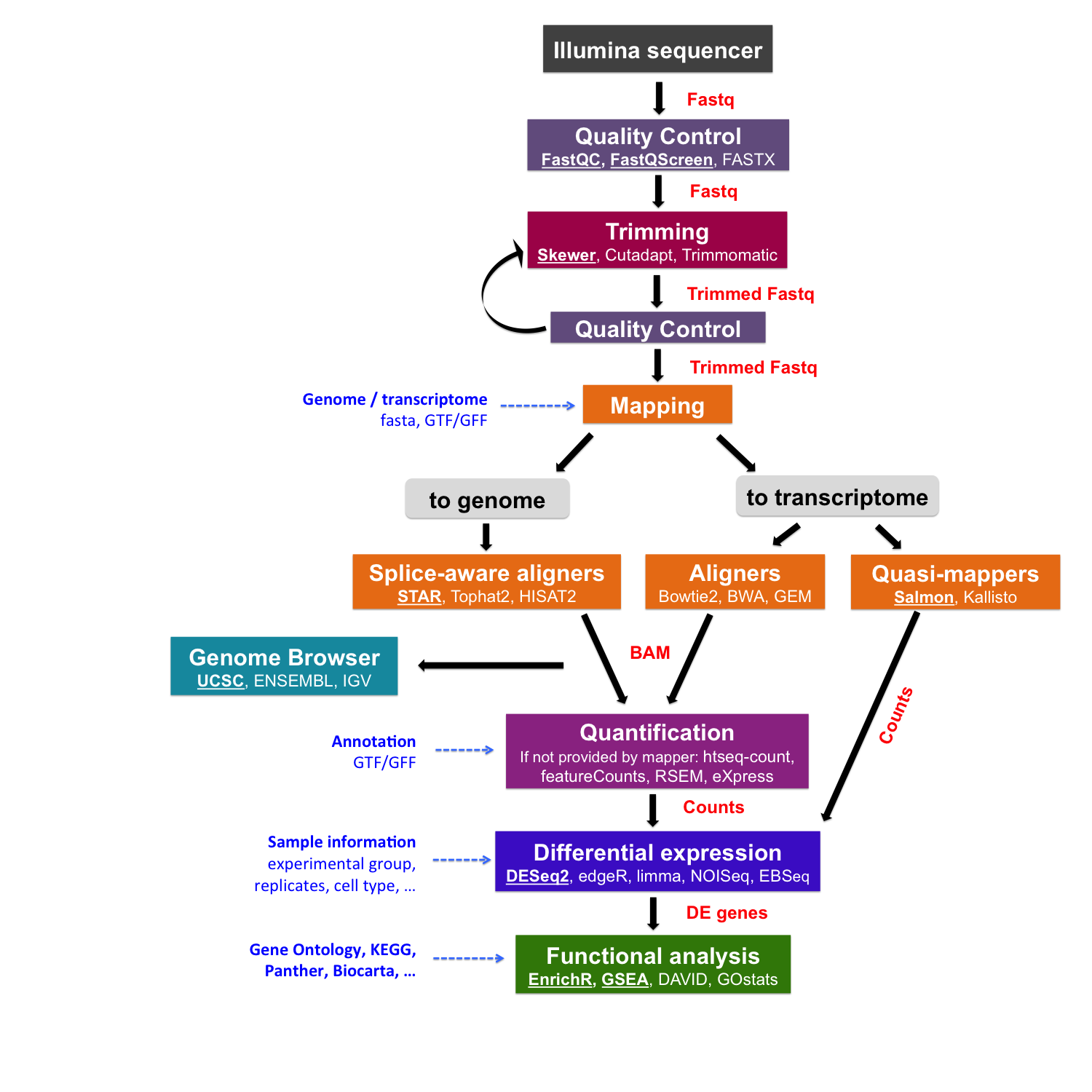
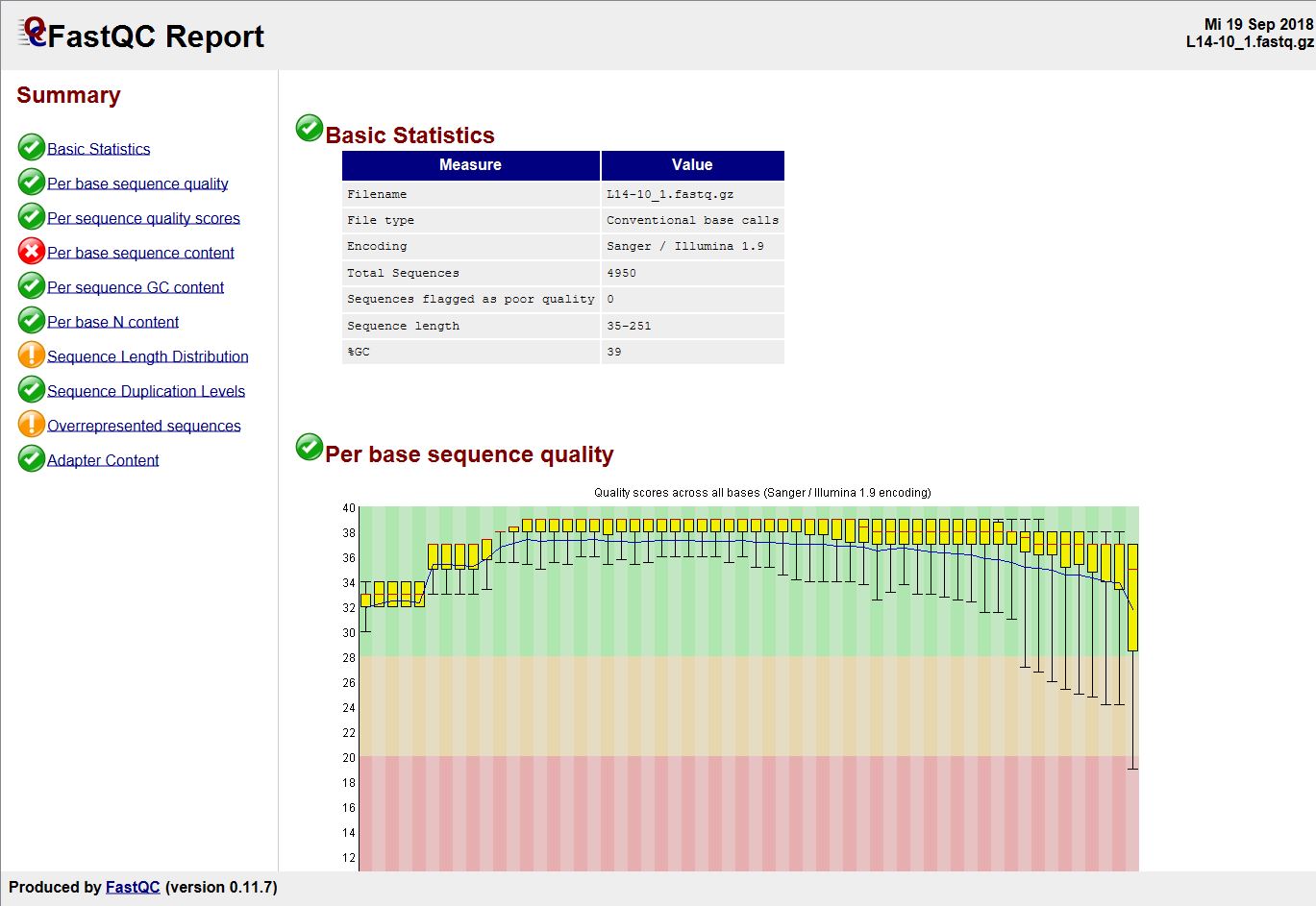
Cutadapt remoYes adapter sequences from high-throughput sequencing reads Introduction Implementation Features

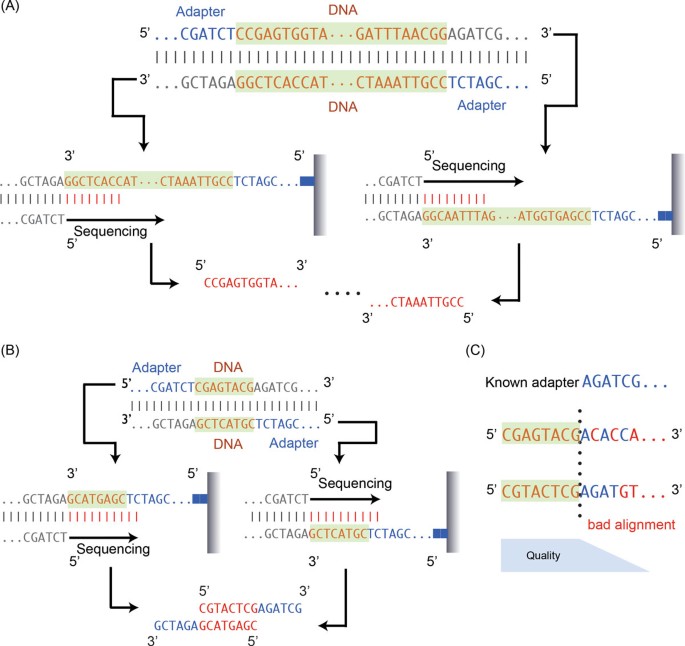
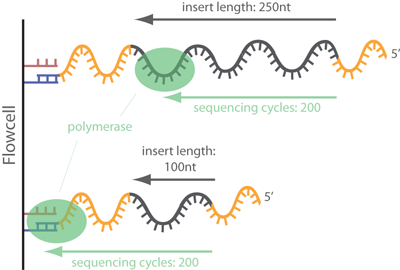
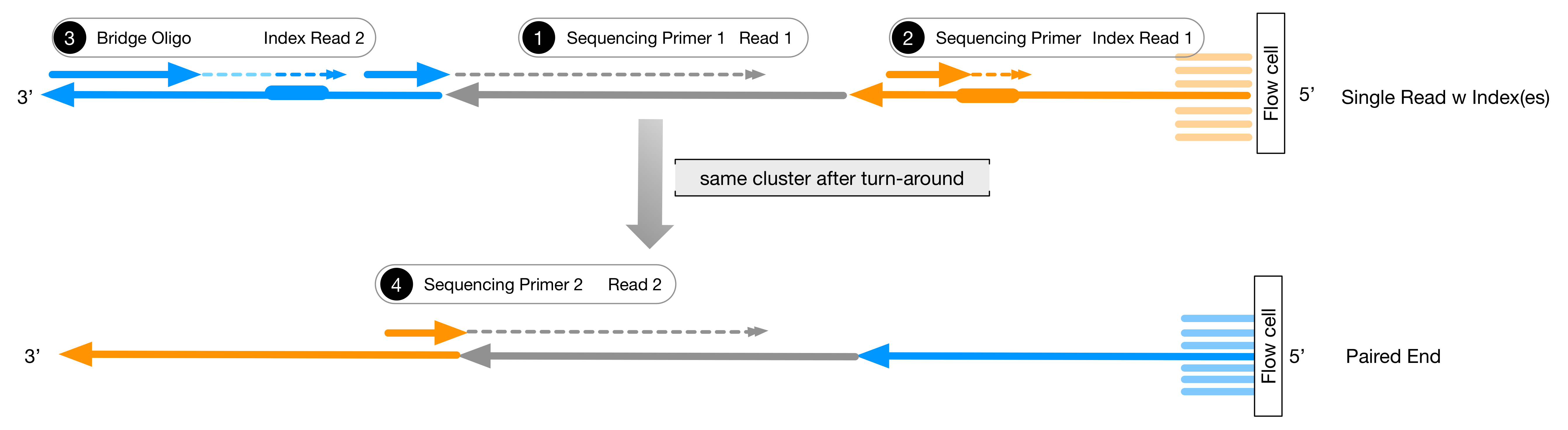
Adapter trimming: Why are adapter sequences trimmed from only the 3' ends of reads - Illumina Knowledge

Adapter trimming: Why are adapter sequences trimmed from only the 3' ends of reads - Illumina Knowledge

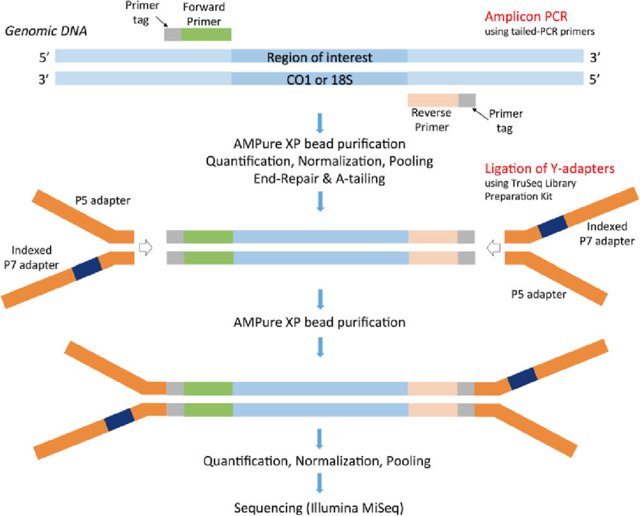
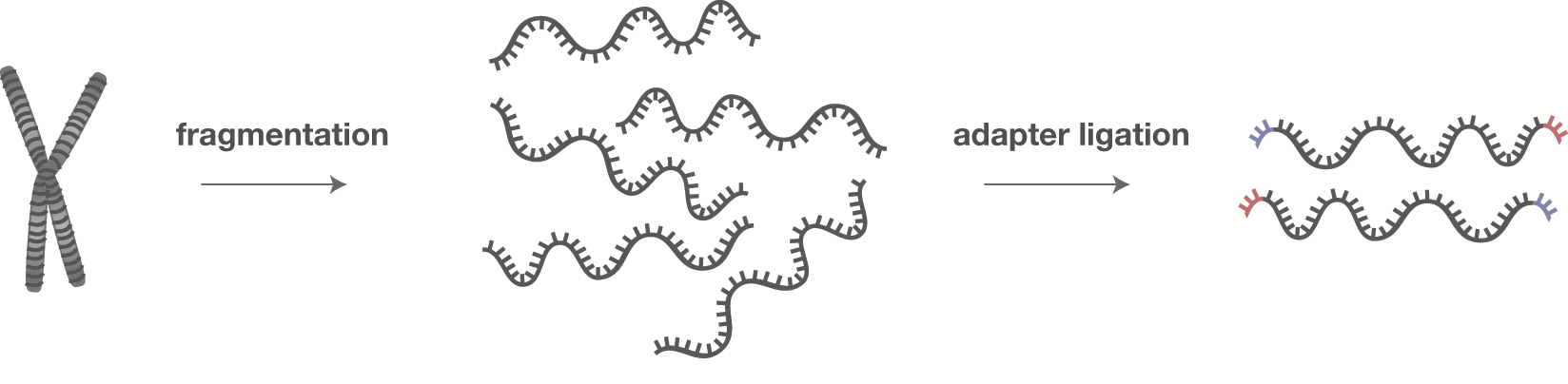
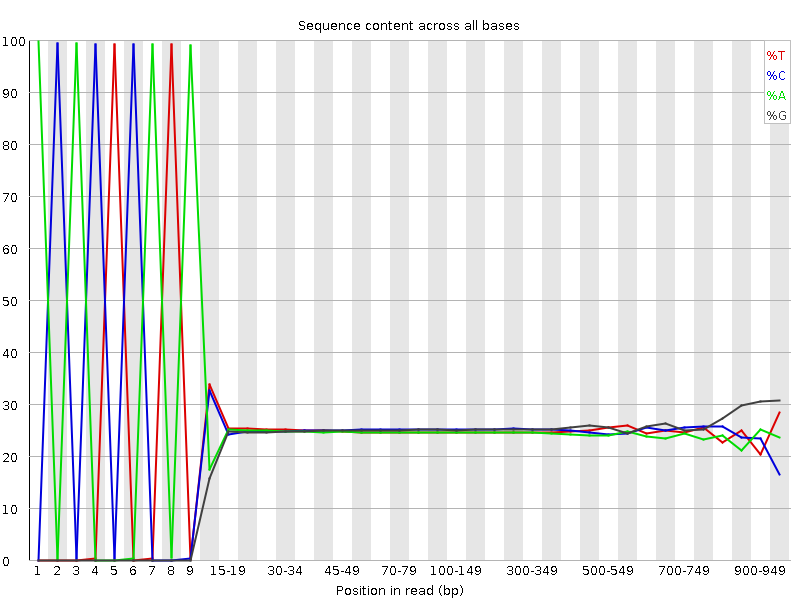
Biomolecules | Free Full-Text | High-Throughput Identification of Adapters in Single-Read Sequencing Data

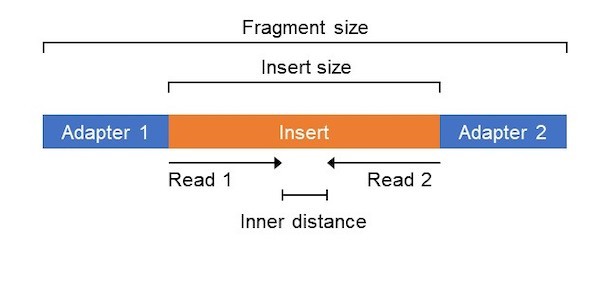
Adapter trimming for single end sRNA-seq data. The reads processed by... | Download Scientific Diagram