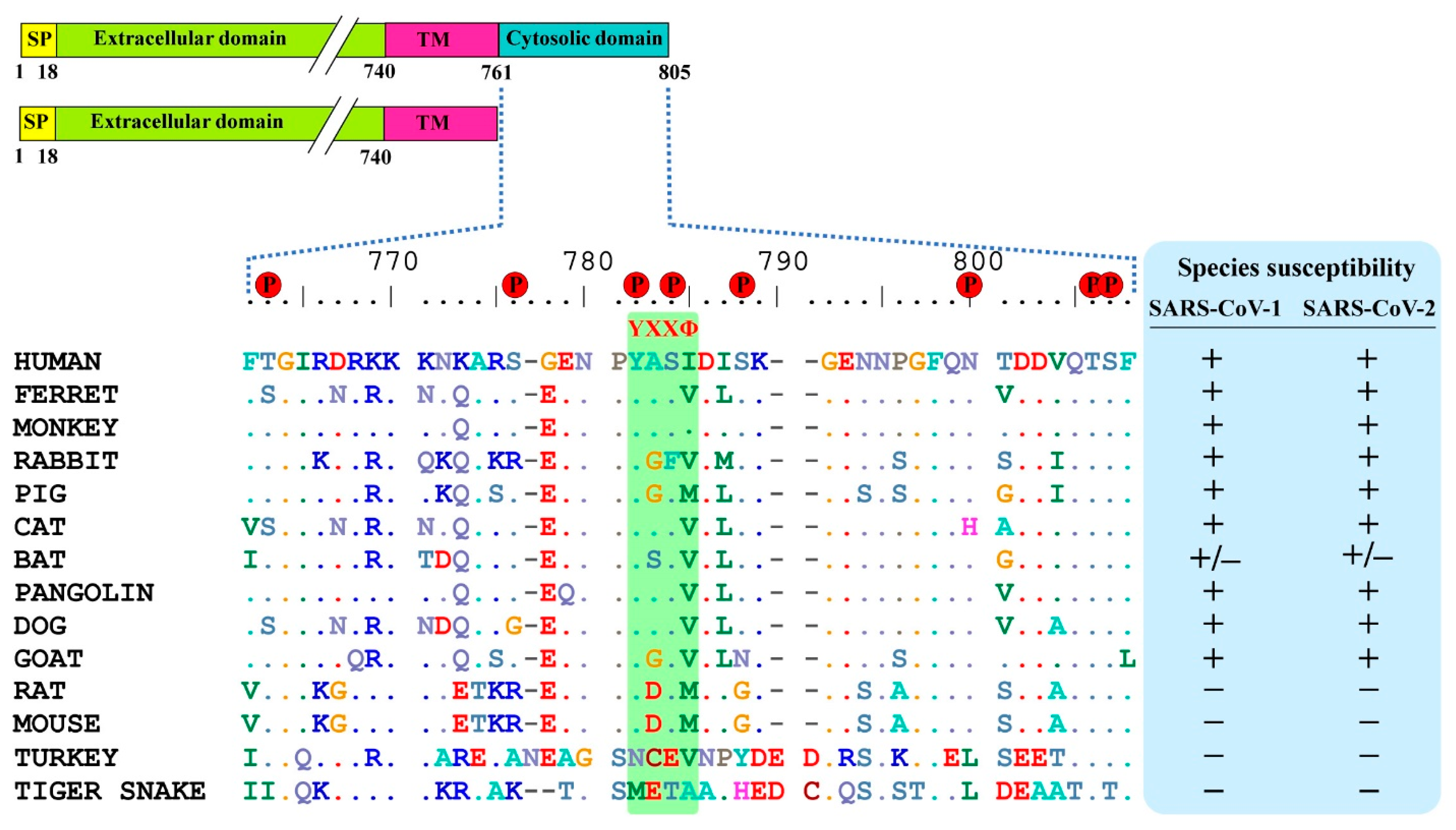
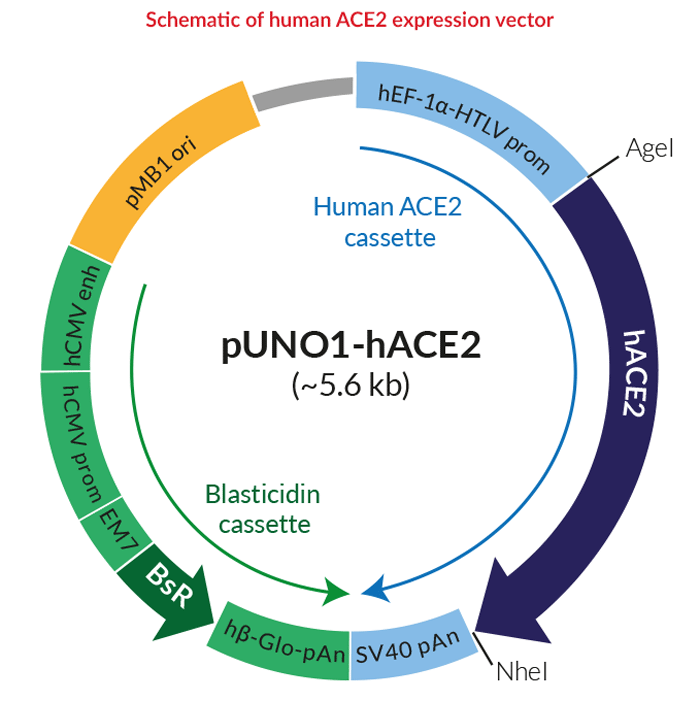
Designed Variants of ACE2-Fc that Decouple Anti-SARS-CoV-2 Activities from Unwanted Cardiovascular Effects | bioRxiv
Frontiers | The Impact of ACE2 Polymorphisms on COVID-19 Disease: Susceptibility, Severity, and Therapy
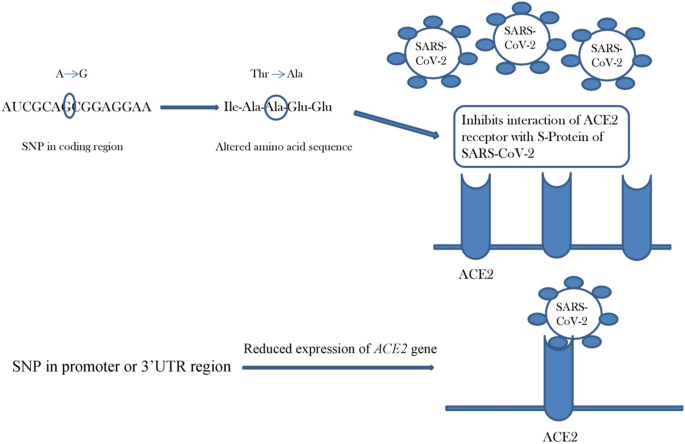
COVID-19 susceptibility: potential of ACE2 polymorphisms | Egyptian Journal of Medical Human Genetics | Full Text

Broad and Differential Animal Angiotensin-Converting Enzyme 2 Receptor Usage by SARS-CoV-2 | Journal of Virology

SARS‐CoV‐2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells | The EMBO Journal

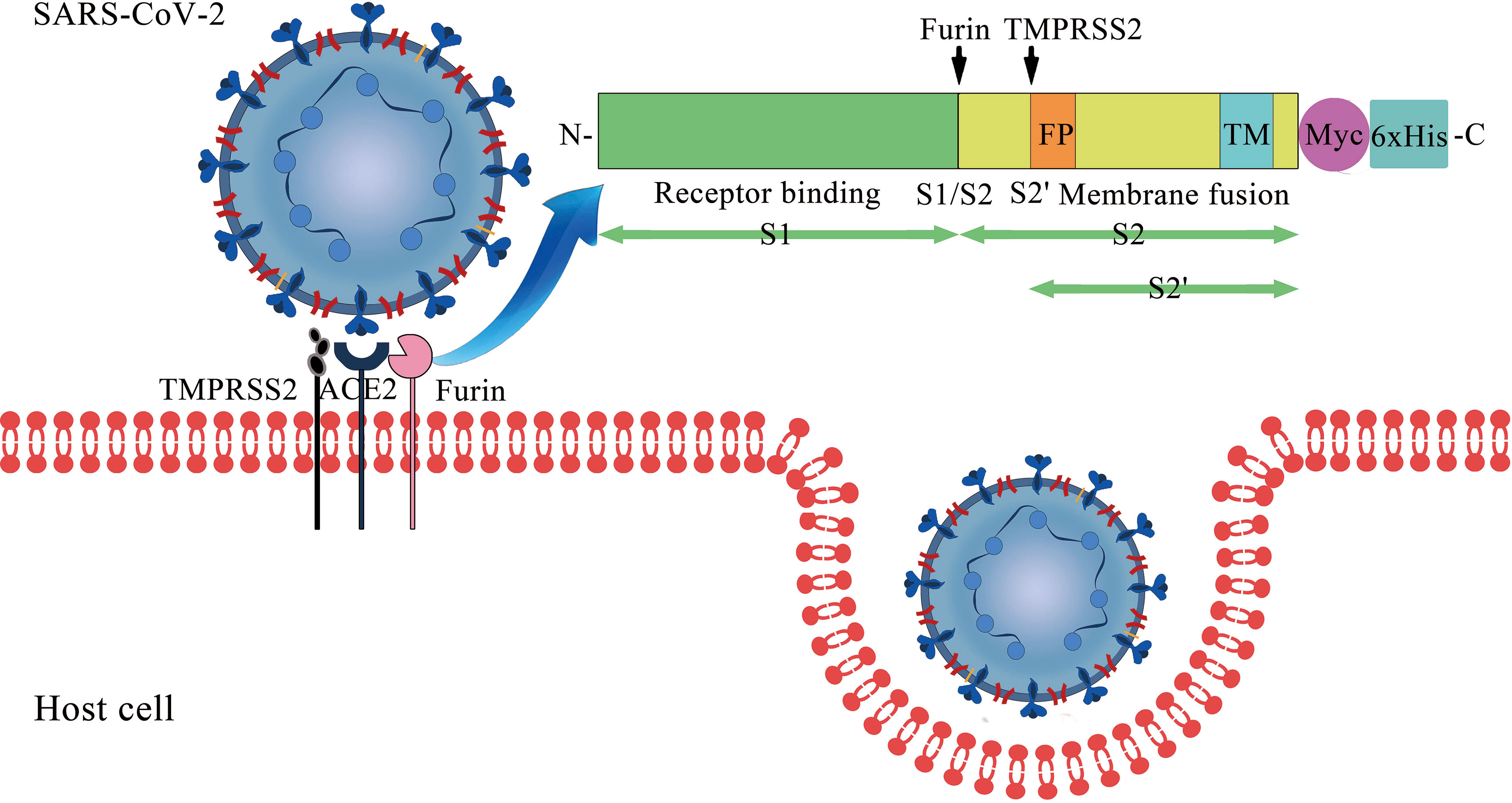
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor - ScienceDirect
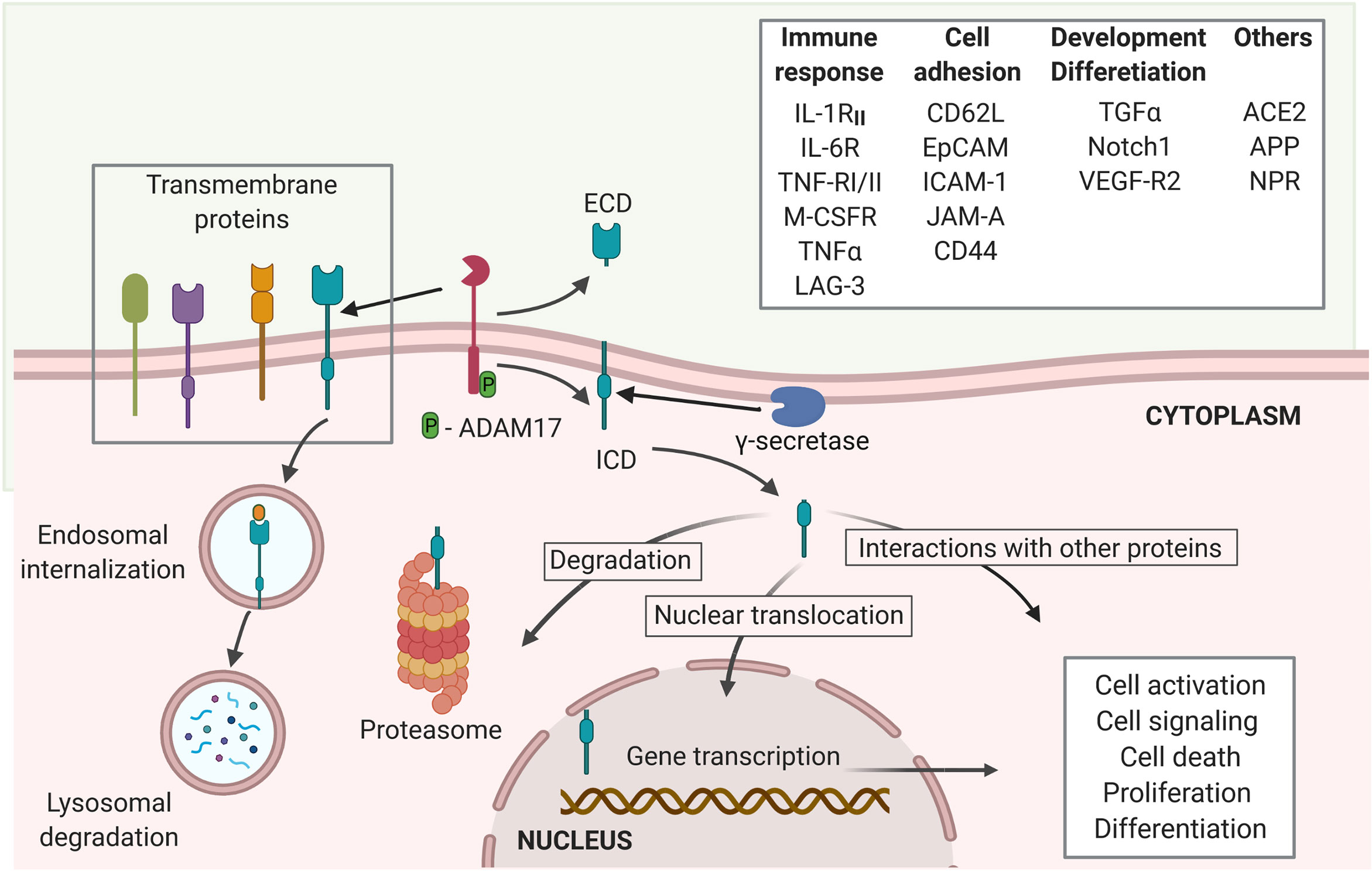
Frontiers | Regulated Intramembrane Proteolysis of ACE2: A Potential Mechanism Contributing to COVID-19 Pathogenesis?
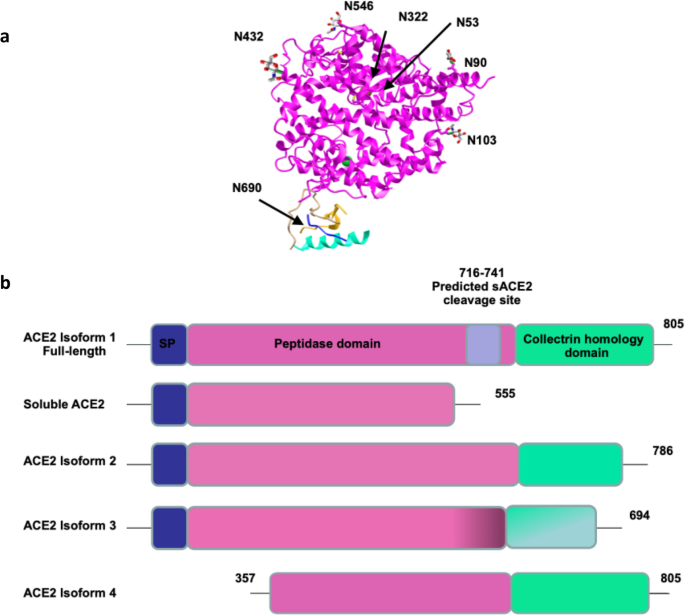
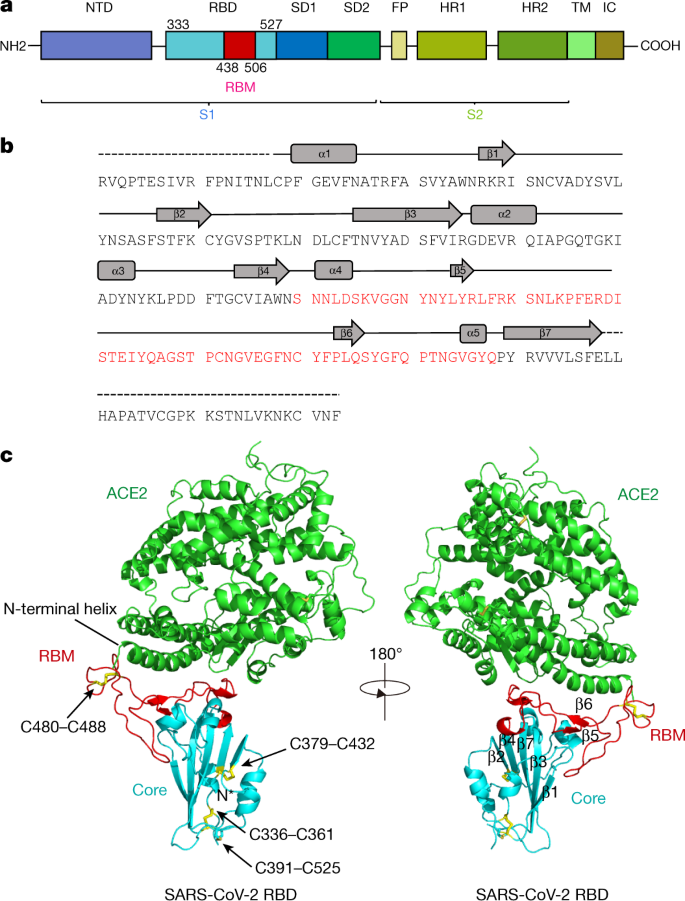
ACE2 Nascence, trafficking, and SARS-CoV-2 pathogenesis: the saga continues | Human Genomics | Full Text
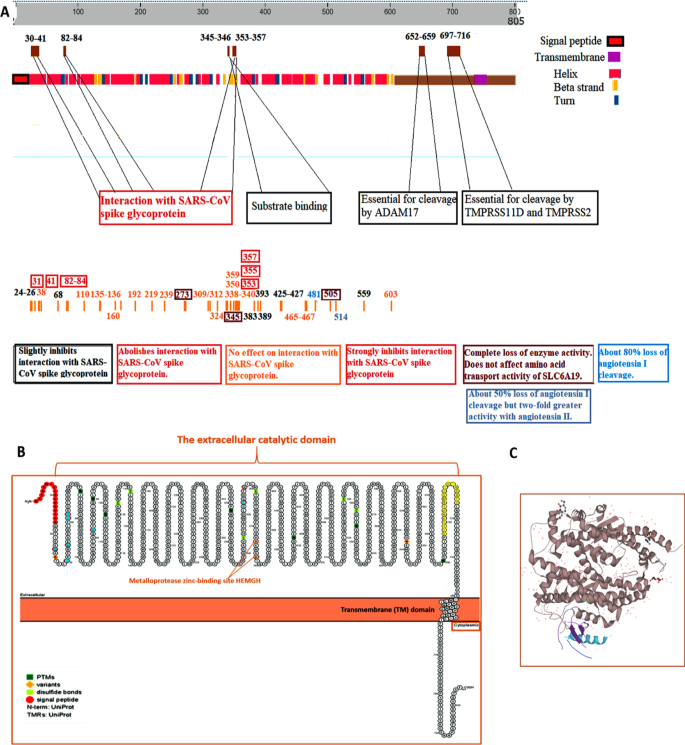
The role of angiotensin-converting enzyme 2 (ACE2) genetic variations in COVID-19 infection: a literature review | Egyptian Journal of Medical Human Genetics | Full Text

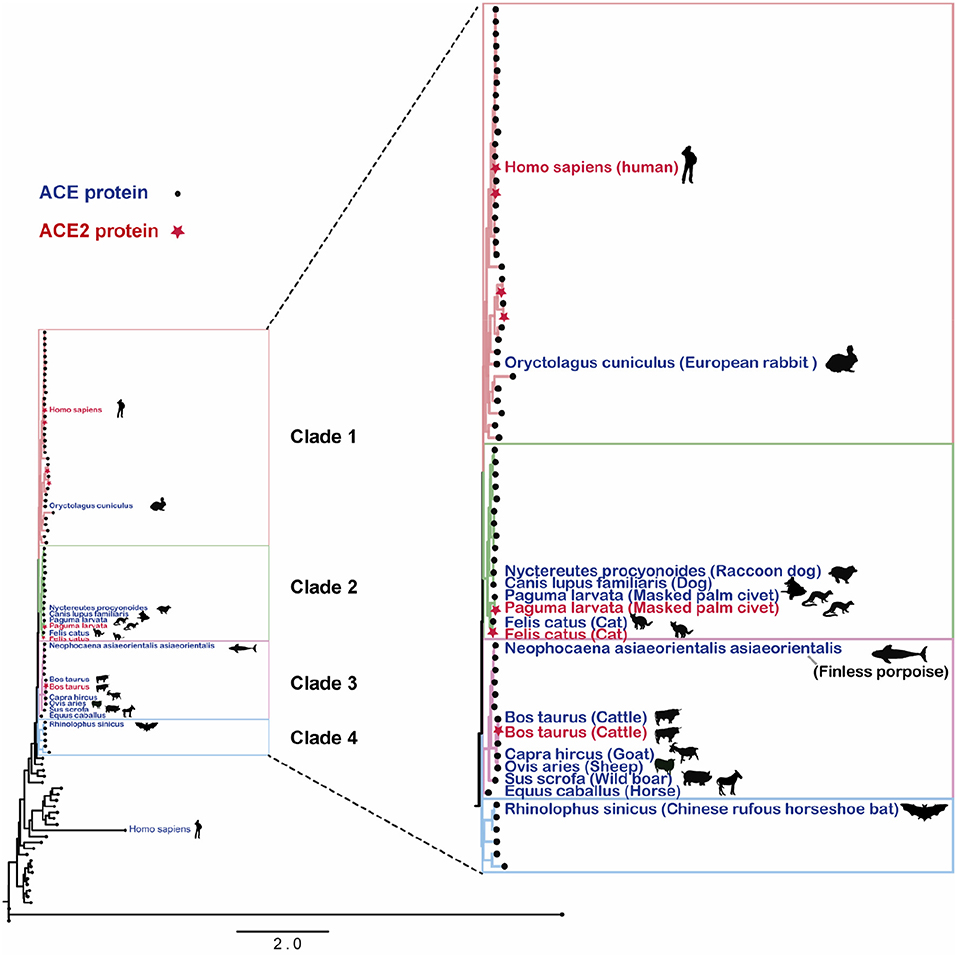
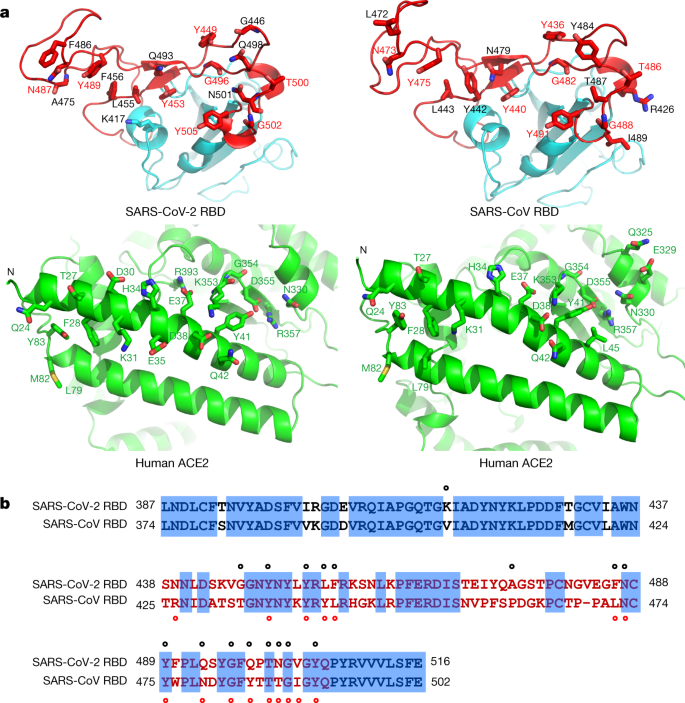
Sequence difference of angiotensin-converting enzyme 2 between nonhuman primates affects its binding-affinity with SARS-CoV-2 S receptor binding domain | Biosafety and Health
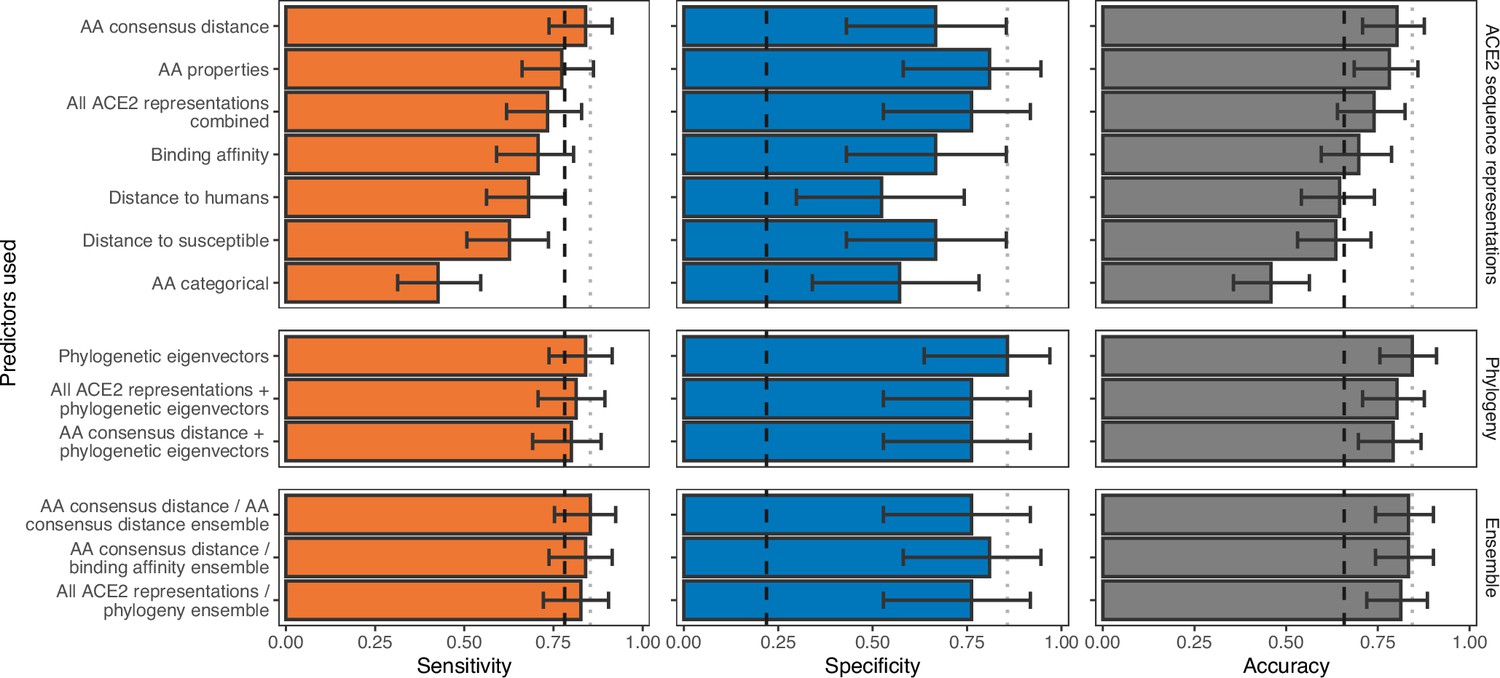
In vitro evolution predicts emerging SARS-CoV-2 mutations with high affinity for ACE2 and cross-species binding | PLOS Pathogens