
General outline of oligo(dT)-based 3′-end sequencing protocols (e.g.,... | Download Scientific Diagram

Poly(A)-ClickSeq – click-chemistry for next-generation 3΄-end sequencing without RNA enrichment or fragmentation | RNA-Seq Blog

TM3'seq: a tagmentation-mediated 3' sequencing approach for improving scalability of RNA-seq experiments | bioRxiv

Multiplex structural variant detection by whole-genome mapping and nanopore sequencing | Scientific Reports
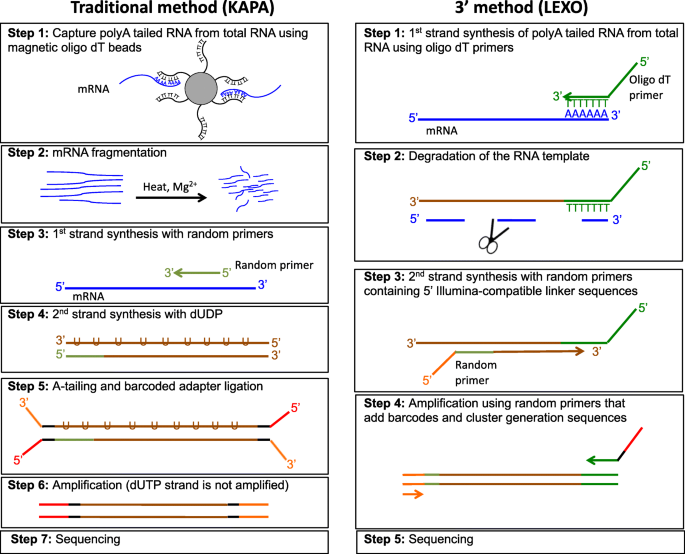
A comparison between whole transcript and 3' RNA sequencing methods using Kapa and Lexogen library preparation methods | BMC Genomics | Full Text

Nano3P-seq: transcriptome-wide analysis of gene expression and tail dynamics using end-capture nanopore cDNA sequencing | Nature Methods
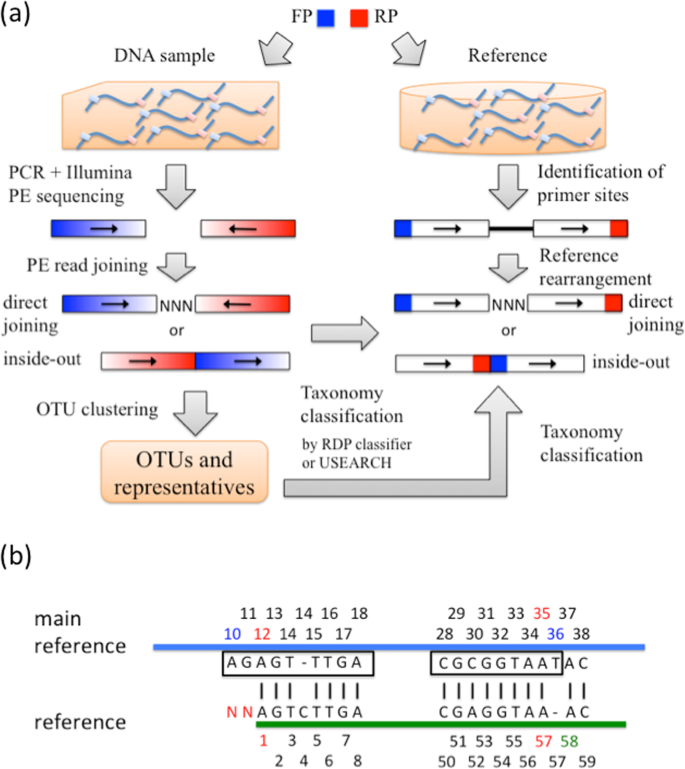
Joining Illumina paired-end reads for classifying phylogenetic marker sequences | BMC Bioinformatics | Full Text

A work flow for 3'end-seq and PacBio Iso-seq. (A) Multiple versions of... | Download Scientific Diagram

3′ end sequencing workflow with oligo(dT) 19 V. a) Overview of the 3′... | Download Scientific Diagram
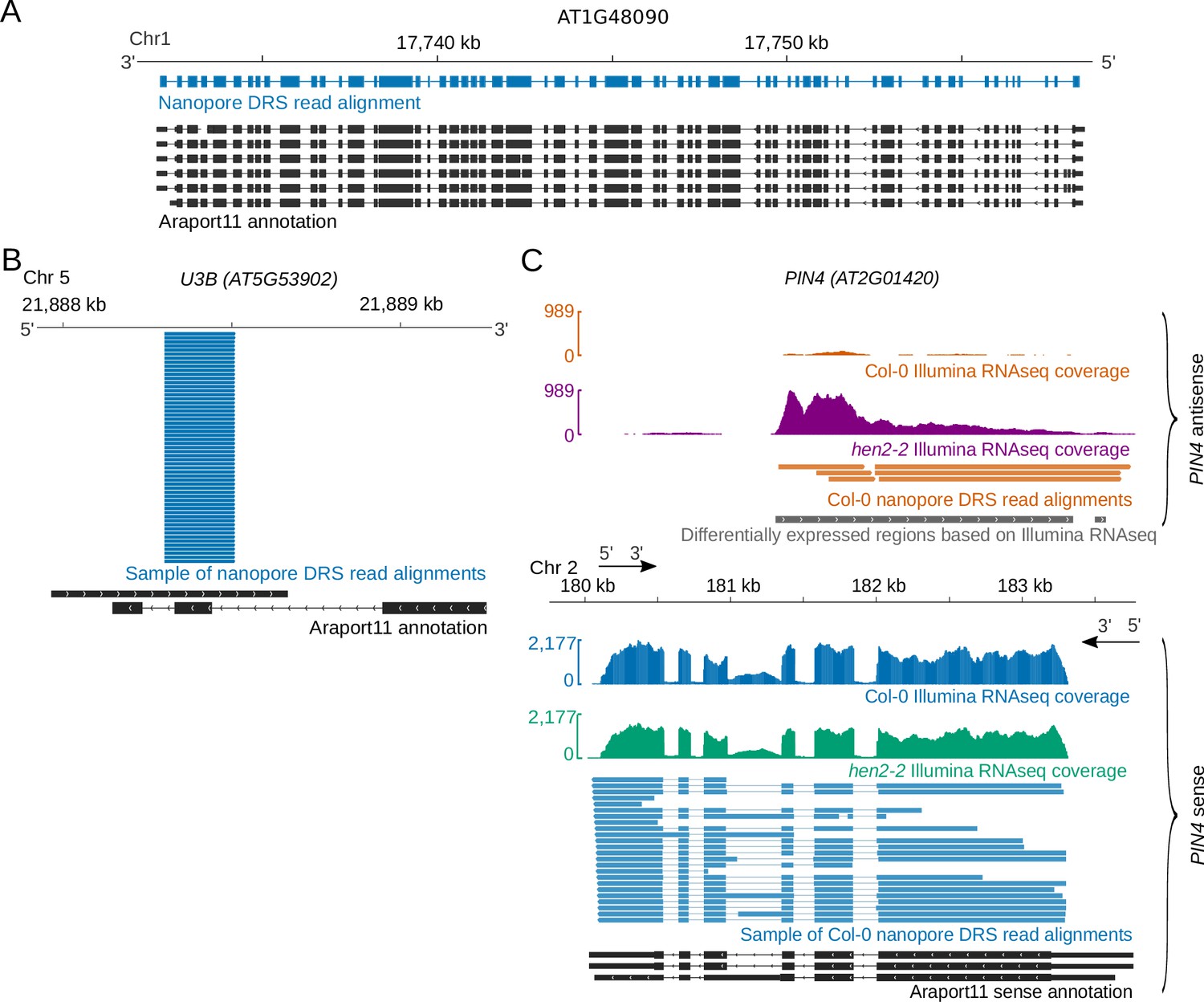

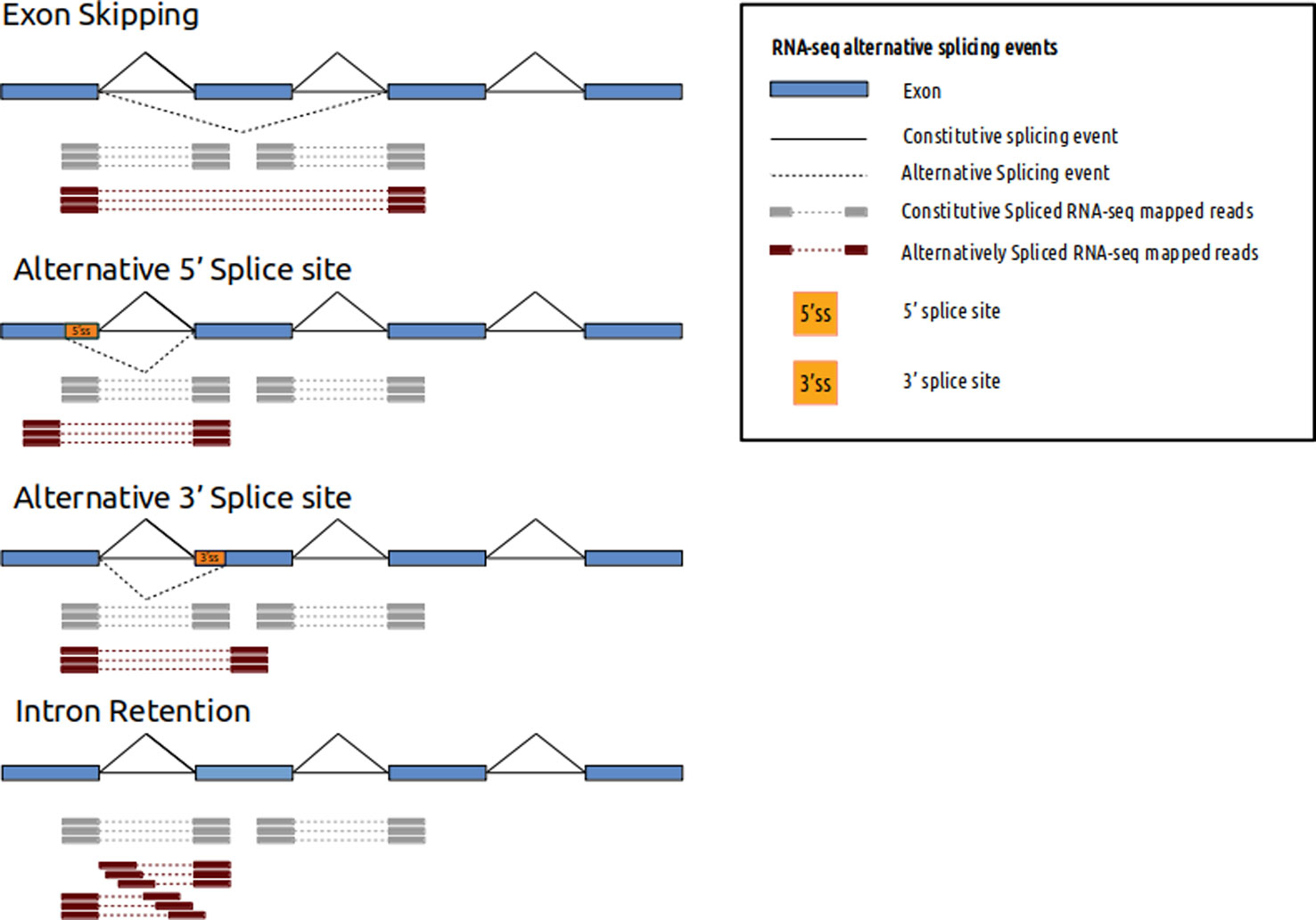
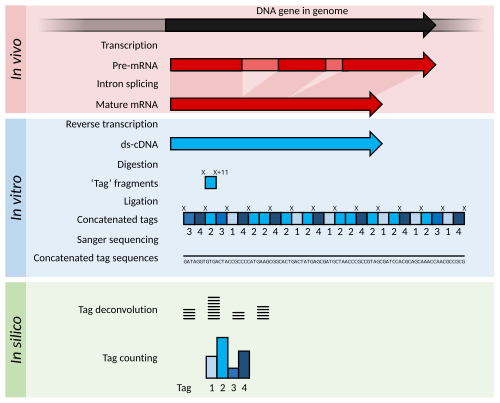





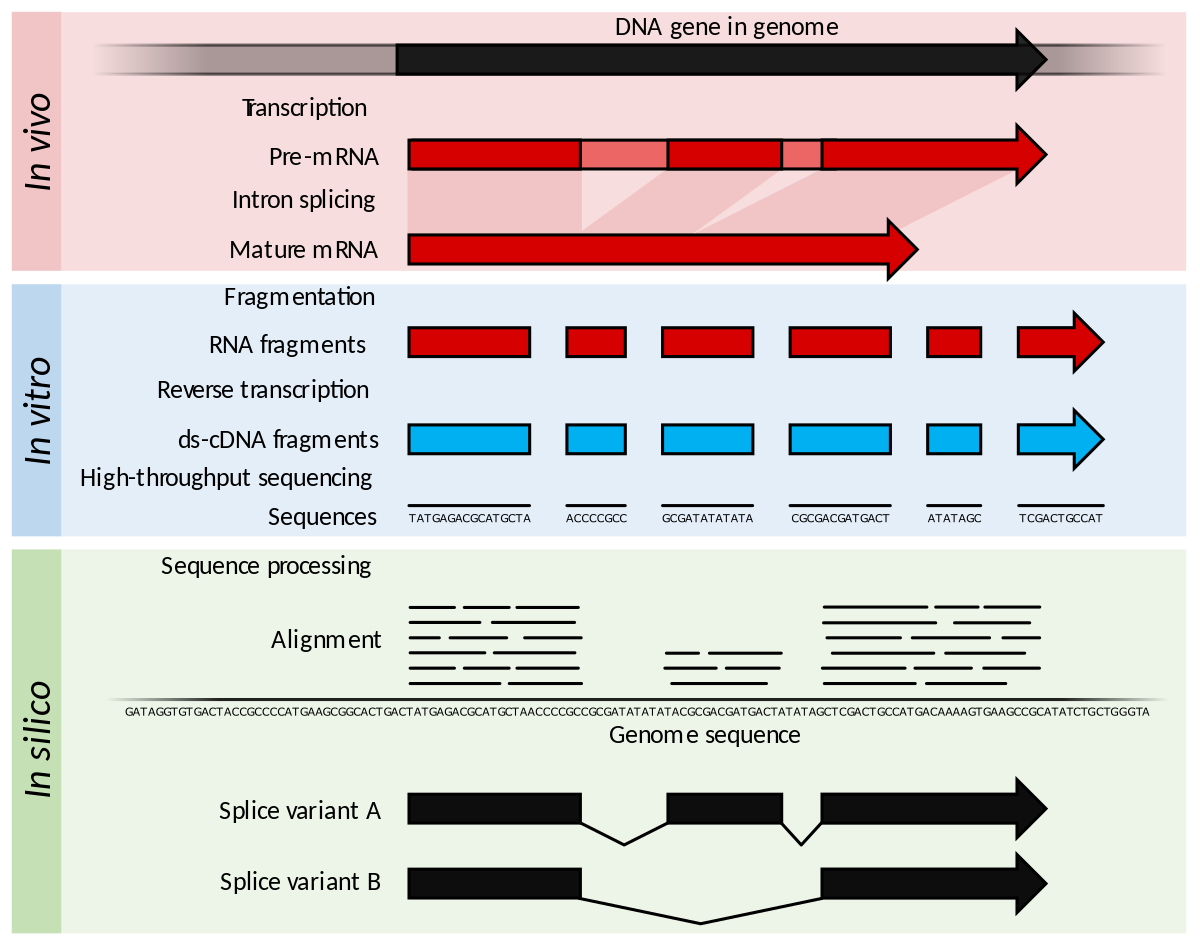


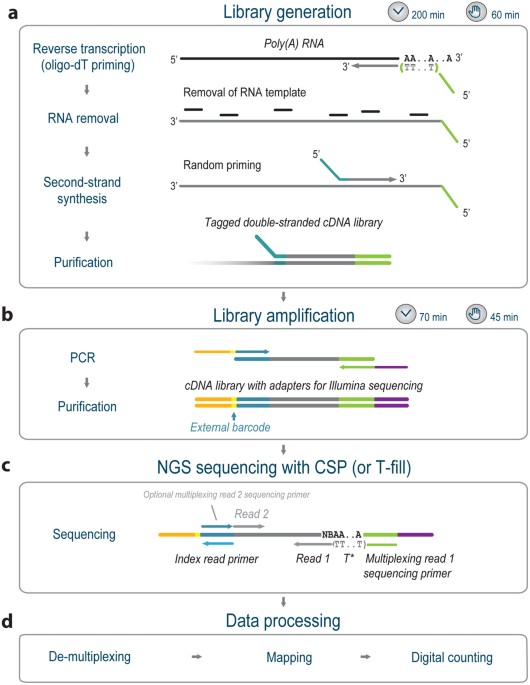



![3′ end sequencing methods. a In DeepSAGe [50] poly(A) + RNAs are... | Download Scientific Diagram 3′ end sequencing methods. a In DeepSAGe [50] poly(A) + RNAs are... | Download Scientific Diagram](https://www.researchgate.net/publication/262339133/figure/fig4/AS:667031334572034@1536044071441/end-sequencing-methods-a-In-DeepSAGe-50-polyA-RNAs-are-captured-by-oligo-dT.png)