Defining the anti-Shine-Dalgarno sequence interaction and quantifying its functional role in regulating translation efficiency | bioRxiv

The possible dual impacts of Shine-Dalgarno (SD) sequences on protein... | Download Scientific Diagram
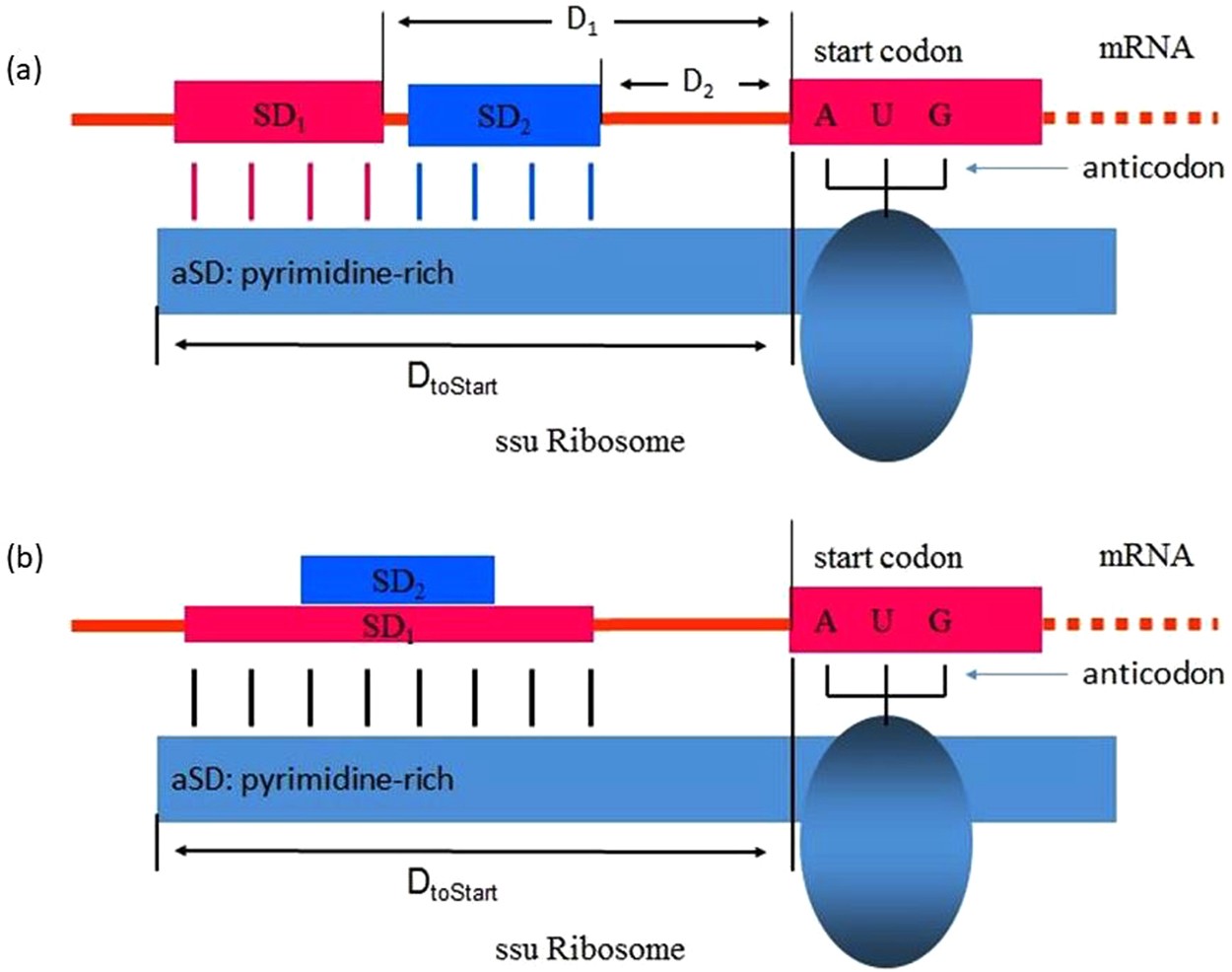
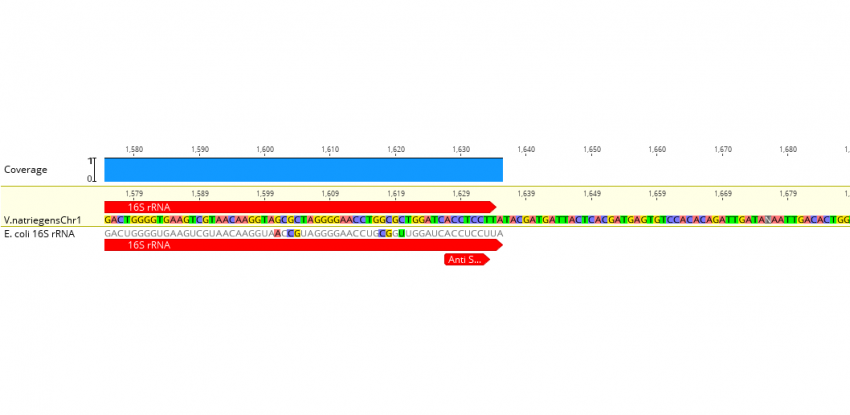
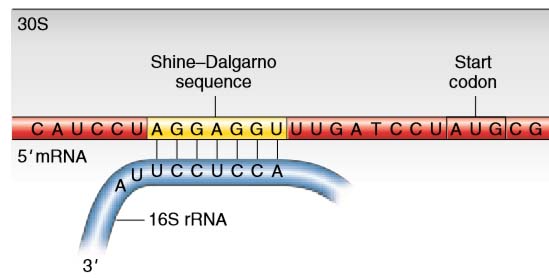
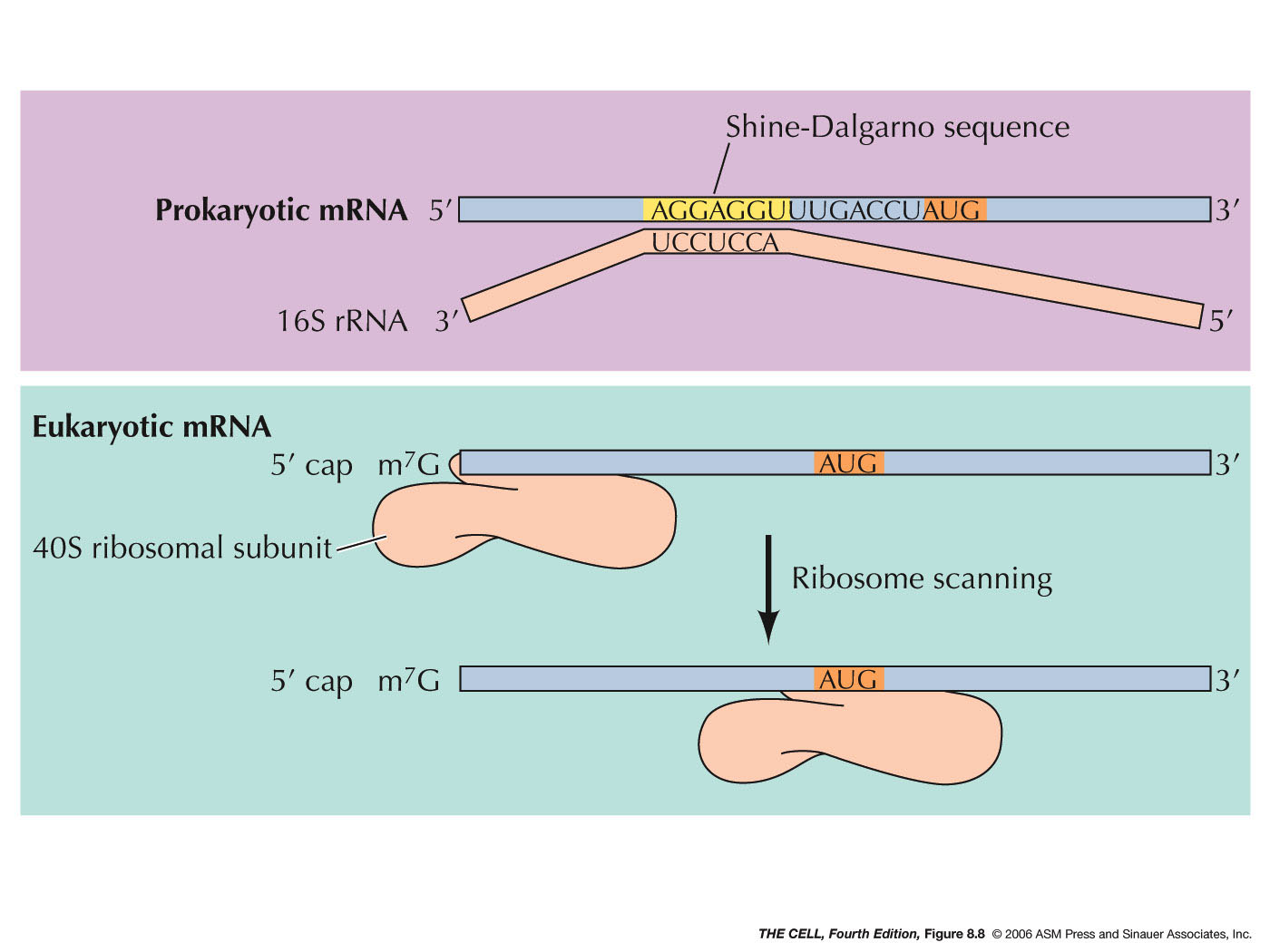
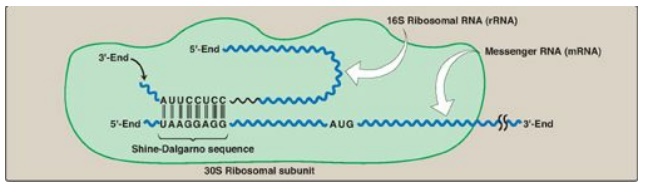
Elucidating the 16S rRNA 3′ boundaries and defining optimal SD/aSD pairing in Escherichia coli and Bacillus subtilis using RNA-Seq data | Scientific Reports

Leveraging genome-wide datasets to quantify the functional role of the anti- Shine–Dalgarno sequence in regulating translation efficiency | Open Biology
![PDF] Re-annotation of 12,495 prokaryotic 16S rRNA 3' ends and analysis of Shine-Dalgarno and anti-Shine-Dalgarno sequences | Semantic Scholar PDF] Re-annotation of 12,495 prokaryotic 16S rRNA 3' ends and analysis of Shine-Dalgarno and anti-Shine-Dalgarno sequences | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/d62ab184b34e5d10344639999cec01b232f895de/4-Figure2-1.png)
PDF] Re-annotation of 12,495 prokaryotic 16S rRNA 3' ends and analysis of Shine-Dalgarno and anti-Shine-Dalgarno sequences | Semantic Scholar
Re-annotation of 12,495 prokaryotic 16S rRNA 3' ends and analysis of Shine- Dalgarno and anti-Shine-Dalgarno sequences | PLOS ONE
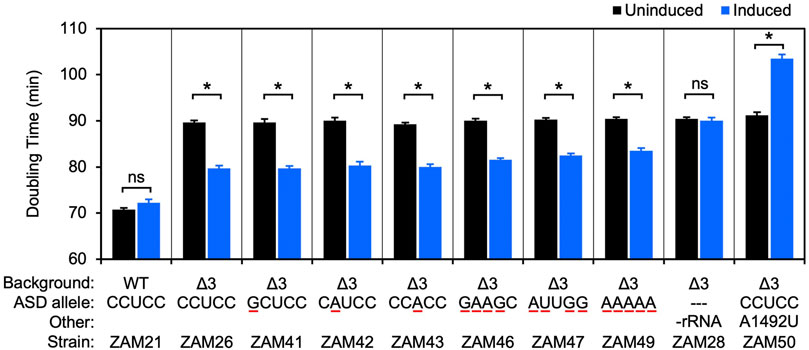
Frontiers | Comparative Analysis of anti-Shine- Dalgarno Function in Flavobacterium johnsoniae and Escherichia coli

An extended Shine–Dalgarno sequence in mRNA functionally bypasses a vital defect in initiator tRNA | PNAS

Extending the Spacing between the Shine–Dalgarno Sequence and P-Site Codon Reduces the Rate of mRNA Translocation - ScienceDirect